2nd Biomedical Visualization Contest Winners

Kitware held its 2nd Annual Contest to find the best biomedical visualization made using the freely-available, open-source Visualization Toolkit and the Insight Registration and Segmentation Toolkit.
Judging for this contest was completed in a secure online environment and the winners were announced at 12th International Conference on Medical Image Computing and Computer Assisted Intervention, held September 20 – 24, 2009 in London, England.
This year’s submissions employed the following software applications in creating their contest submissions: caBIG-XIP, 3D Slicer, Osirix, MedINRIA, IRCAD’s VR-Render, and MITK. Each of these applications were built on top of Kitware’s VTK and/or ITK open source toolkits.
Prizes and awards:
- The first place award was $750 and a Kitware polo shirt
- The second place award was $250 and a Kitware polo shirt
To view all 20 contest entries, please visit the contest website. And the winners are…
Second Place
Brain Connectivity as Network Visualization
Stephan Gerhard, Patric Hagmann, EPFL, Lausanne
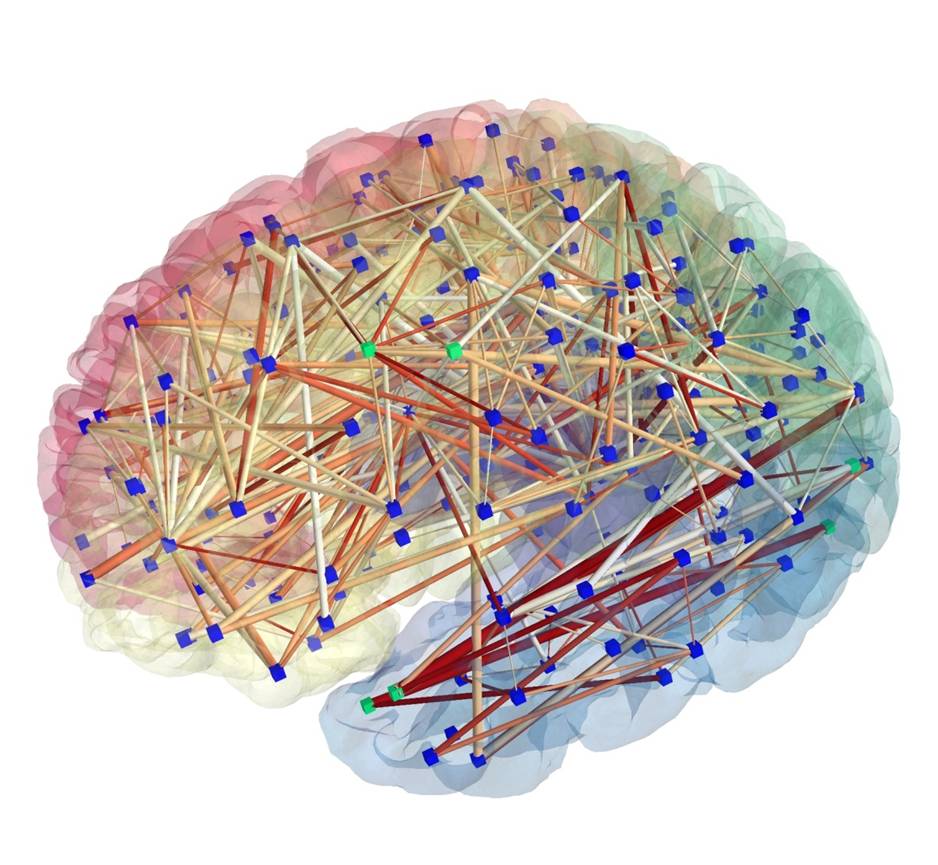
Image Description: Diffusion Spectrum Imaging data has been processed by the Connectome Mapping Pipeline, bundling white matter tracts and producing networks in multiple resolutions (number of nodes) and different edge attributes. The image was rendered using the ConnectomeViewer application. This network has 258 nodes corresponding to cortical and subcortical ROIs (shown as blue cubes and as transparent surfaces), the edges depict threshold and color-coded densities between connecting ROIs. Green cubes are picked for further processing with the integrated Python interface. See also http://www.connectome.ch/
Role of Open Source: The ConnectomeViewer application’s framework was built using Enthought Envisage, with Mayavi2 using TVTK as plugin. TVTK wraps VTK to trait-enable Python. Nifti, Gifti, Connectome File Format (CFF) were also employed.
First Place
Computer Assisted surgery of pancreas using Augmented Reality
Luc Soler and Jacques Marescaux; IRCAD, France
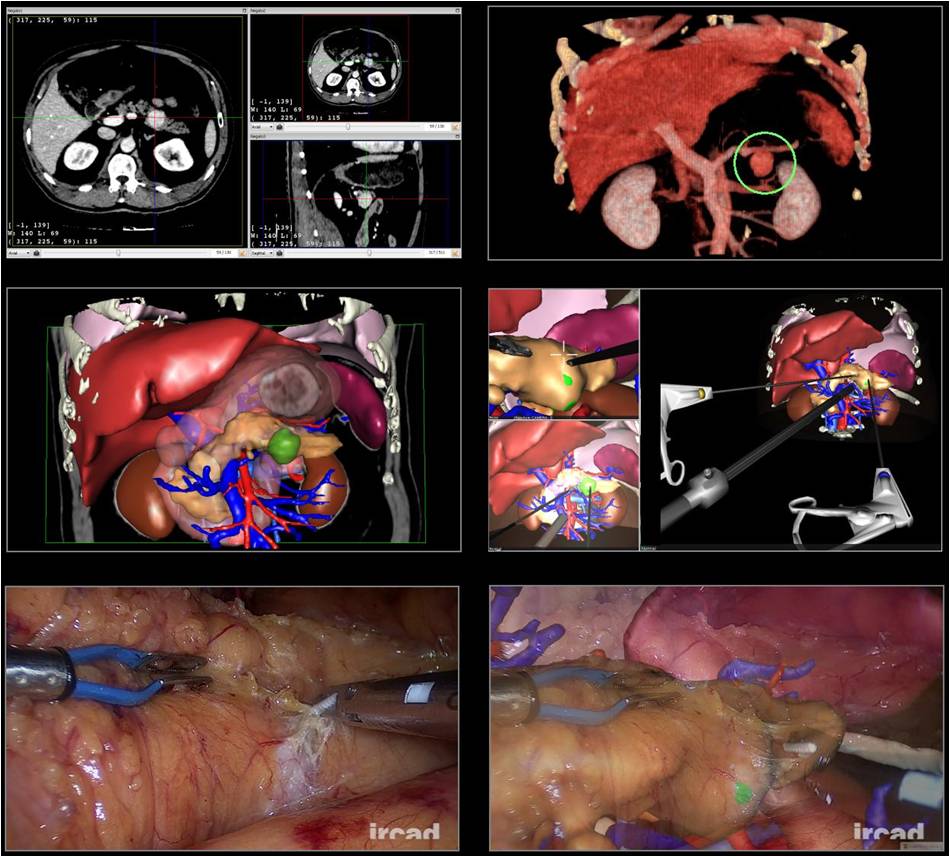
Image Description: Vascular anomaly and pulmonary sequestration diagnosis using IRCAD software applications on a thoracic CT-scan of a 1 month old baby with direct volume rendering (first line) and surface rendering resulting from organ segmentations (second line) in anterior view (left) and posterior view (right). The resulting preoperative surgical planning allows to reach arteries and the lung area to be cut with an optimal back intercostal tool positioning that is perfectly reproduced intraoperatively (last line).
Role of Open Source: Image segmentation is done using ITK (morphological mathematics, topological, geometrical and textural operators) included in IRCAD 3DVPM software. VTK is used by IRCAD VR-Render software (www.ircad.fr/software) for direct volume rendering (3D texturing with vtkVolumeTextureMapper3D). These software applications work with other Open Source systems like wxWidgets (for GUI), gdcm/dcmtk (for loading DICOM), vtkinria3d, vgSDK or boost.
Kitware would like to thank the following community members for helping to judge this contest:
- Nicholas Ayache, INRIA, Sophia-Antipolis
- J. Michael Fitzpatrick, Vanderbilt University
- Alejandro Frangi, Pompeu Fabra University
- Henkjan Huisman, Radboud University Medical Centre Nijmegen
- Nico Karssemeijer, Radboud University Medical Centre Nijmegen
- Dean Inglis, McMaster University
- Marc Niethammer, The University of North Carolina
- Steve Pieper, Isomics, Inc.
- Julia Schnabel, University of Oxford
- Martin Styner, The University of North Carolina
- Debora Testi, BioComputing Compentence Centre
- Marco Viceconti, Istituto Ortopedico Rizzoli
Kitware would like to thank the following community members for helping to make this contest possible:
- Stephen Aylward
- Julien Jomier
- Steve Pieper
- Patrick Reynolds
- Daniel Rueckert
- Betty Yue