DeepAtlas in MONAI: Jointly Learning Registration and Segmentation
Introduction
Image segmentation and image registration play an important role in many applications of medical image analysis. Segmentation locates and defines the precise regions occupied by anatomical structures, allowing for the analysis of their size and shape and enabling other algorithms to restrict their focus to specific regions. Registration aligns the anatomical features between two images, allowing for a meaningful comparison of structures and providing a common coordinate system in which to carry out further analysis.
Deep learning has rapidly advanced the state of the art in our approaches to both of these problems. Given enough data, complex segmentation models can now be trained in an out-of-the-box fashion with almost no need for domain knowledge. Extremely fast registration models can be trained on problem-specific image distributions.
However, getting a large amount of labeled training data can be a real challenge in the world of medical imaging. Manual segmentation requires experts, and it is often laborious because the images are three dimensional. This makes segmentation data quite expensive. Hence we often find ourselves with an abundance of images and a shortage of segmentation labels. The DeepAtlas approach [2] was designed by our collaborators at the University of North Carolina Biomedical Image Analysis Group to tackle this type of situation. In this post, we provide an overview of our implementation of this methodology in the MONAI framework [5].
DeepAtlas
The DeepAtlas approach solves the challenge of limited segmentation data by jointly learning networks for image registration and image segmentation tasks. To see how it works, consider that registration and segmentation are closely related:
- Segmentation can be used to evaluate the performance of a registration algorithm. A good registration algorithm should align anatomical structures, and therefore it should align segmentations.
- Registration can be used to produce segmentations. Given one image– an atlas— with segmentation information, any other image can be segmented by simply registering it to the atlas.
DeepAtlas leverages these ideas and turns them into a training program, where segmentation and registration models act as a form of weak supervision for each other:
- Instead of merely being used to evaluate a registration model, a segmentation model can provide an additional objective to help train a registration model. We call this objective the anatomy loss.
- Using just a few manually created segmentations as atlases, a registration model can be used to generate new segmentations, essentially providing a source of data augmentation for segmentation model training.
By applying these strategies in alternation, we can train a registration model and a segmentation model together, allowing them to build each other up even when there is very little ground truth segmentation data available. This can unleash a potentially massive collection of unlabeled images as useful training data for segmentation.
Implementation in MONAI
MONAI is an open source, community-supported, PyTorch-based framework for deep learning in healthcare imaging, useful for rapidly developing and deploying deep learning models. We have created and made available a tutorial notebook that demonstrates the DeepAtlas methodology for segmentation of brain structures from MRI images. The tutorial is well documented for machine learning developers and practitioners to apply the methodology in their own research.
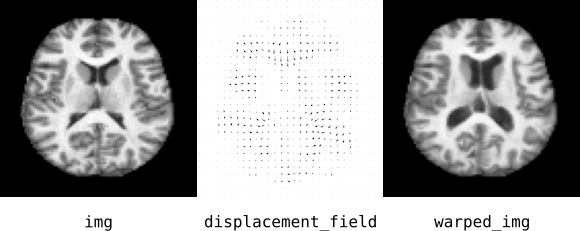
MONAI provides components that make it easy to work with image registration concepts. Suppose you have an image, img, and a displacement vector field, displacement_field. To warp the image along the vector field, use the Warp block:
warp = monai.networks.blocks.warp.Warp(mode='bilinear')
warped_img = warp(img, displacement_field)
You can also warp segmentation labels in the same way. Suppose you have a registration network reg_net and a segmentation network seg_net. Starting from two images img1 and img2, here is an example where we compute the anatomy loss:
# do registration
displacement_field = reg_net(torch.cat((img1,img2), dim=1))
# do segmentation
seg1 = seg_net(img1).softmax(dim=1)
seg2 = seg_net(img2).softmax(dim=1)
# warp segmentation using the same warp block defined above
seg2_warped = warp(seg2, displacement_field)
# compute multiclass dice loss
dice_loss = monai.losses.DiceLoss()
anatomy_loss = dice_loss(seg2_warped, seg1)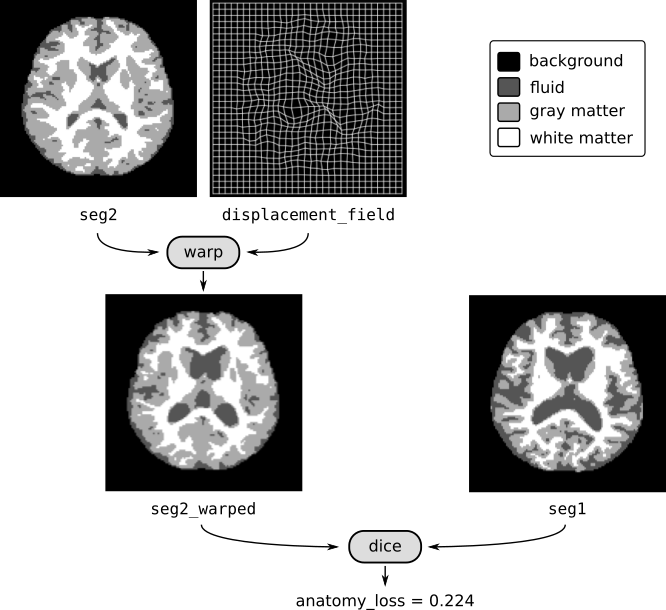
In the tutorial notebook, we work with the OASIS-1 dataset to construct registration and segmentation models for brain MRIs. The figure above shows an axial projection of the 3D images.
For our examples, the registration network is a fully convolutional neural network that directly produces a displacement vector field. This is just one way to do non-parametric registration; one can replace this approach by any function that maps a pair of images to a displacement field (as long the gradients are there for training).
With our simple displacement field approach, training the registration network based on anatomy loss or image similarity alone does not work in practice. We need some form of regularization; otherwise, the network learns to produce wild and physically unrealistic deformations just to get a pixel-perfect match between images. For this we use bending energy loss. Designing the training objective function for registration becomes a balancing act: on one hand we want deformations to match up image structures, and on the other hand we want deformations to be reasonably tame.
In the notebook, we have included a visualization of folds to evaluate model performance and to aid the process of choosing regularization hyperparameters. Folds are image locations where the Jacobian determinant of a deformation takes negative values. Looking out for these is one way to discern how bad our deformations are. It doesn’t tell us the whole story, but it can be useful for tuning the objective function. Below is a colormap of the Jacobian determinant of the registration deformation shown above, with folds shown in red.
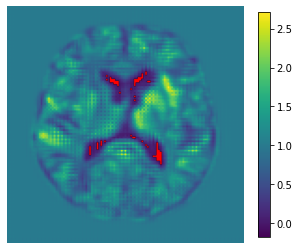
Observing a lot of folds tells us that we may want to turn up the regularization, introduce an inverse consistency loss [1], or change the registration paradigm entirely to one that is more inherently smooth [3, 4].
Conclusion
In this post, we have presented our implementation of the DeepAltas methodology in MONAI. Having an open access implementation of this methodology in MONAI makes it easier for other researchers and machine learning engineers to apply it in their projects. We are using DeepAtlas in an NIH funded research project, where we are developing morphometric analysis techniques to investigate local differences in brain anatomy. Our goal is to enable clinical researchers to discover neurological effects related to disease progression.
About Kitware Inc. Kitware provides software and R&D services for customers looking to build medical image deep learning products and processes using MONAI and other deep learning frameworks. Kitware collaborates with customers to solve the world’s most complex scientific challenges through customized software solutions. Kitware delivers innovation by focusing on advanced technical computing, state-of-the-art artificial intelligence, and tailored software solutions. Since its founding in 1998, Kitware has developed a reputation for deep customer understanding and technical expertise, honest interactions, and open innovation.
Acknowledgment
This work was supported by the National Institutes of Health under Award Number R42MH118845. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
[1] Hastings Greer, Roland Kwitt, François-Xavier Vialard, and Marc Niethammer. ICON: learning regular maps through inverse consistency. CoRR, abs/2105.04459, 2021. [ arXiv ]
[2] Zhenlin Xu and Marc Niethammer. Deepatlas: Joint semi-supervised learning of image registration and segmentation. In Dinggang Shen, Tianming Liu, Terry M. Peters, Lawrence H. Staib, Caroline Essert, Sean Zhou, Pew-Thian Yap, and Ali Khan, editors, Medical Image Computing and Computer Assisted Intervention — MICCAI 2019, pages 420–429, Cham, 2019. Springer International Publishing. [ arXiv | DOI ]
[3] Xiao Yang, Roland Kwitt, Martin Styner, and Marc Niethammer. Quicksilver: Fast predictive image registration – a deep learning approach. NeuroImage, 158:378–396, 2017. [ arXiv | DOI ]
[4] Zhengyang Shen, Xu Han, Zhenlin Xu, and Marc Niethammer. Networks for joint affine and non-parametric image registration. In 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pages 4219–4228, 2019. [ arXiv | DOI ]
[5] MONAI Consortium. MONAI: Medical Open Network for AI, 3 2020. [ DOI | http ]