Faster AI deployment in 3D Slicer with the nnU-Net Extension
Kitware is proud to present the Slicer nnU-Net extension. This extension permits quick testing and deployment of nnU-Net-based segmentation models in 3D Slicer.

nnU-Net is a segmentation library that is the backbone to several very popular medical machine learning segmentation models such as the Total Segmentator and the Dental Segmentator. The nnU-Net library is a fully automated, data-driven, pipeline allowing to very quickly reach state-of-the-art performances on multi-class segmentation datasets.
The Slicer NNUNet extension was created to be able to quickly install the nnUNet library and its dependencies into 3D Slicer, and to run a segmentation given only the nnUNet’s trained model.
Using the extension
The extension can be installed directly from Slicer’s Extension Manager starting preview version 5.7 onward.
When using the first time the extension, its dependencies can be installed using the nnUNet Install section.

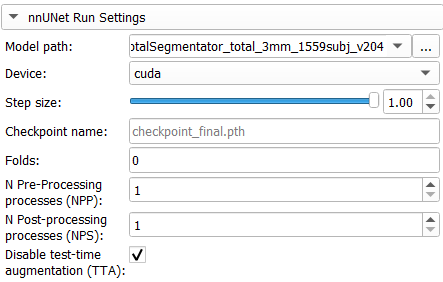
Once the nnUNet is correctly installed, click on the nnUNet Run Settings button to set the path to the Model to use. This path will be saved for further usage after the first segmentation.
The provided model path should contain the nnUNet dataset.json file. To test this extension, you can download the Total Segmentator NNUNet weights and use the CTChest Sample.
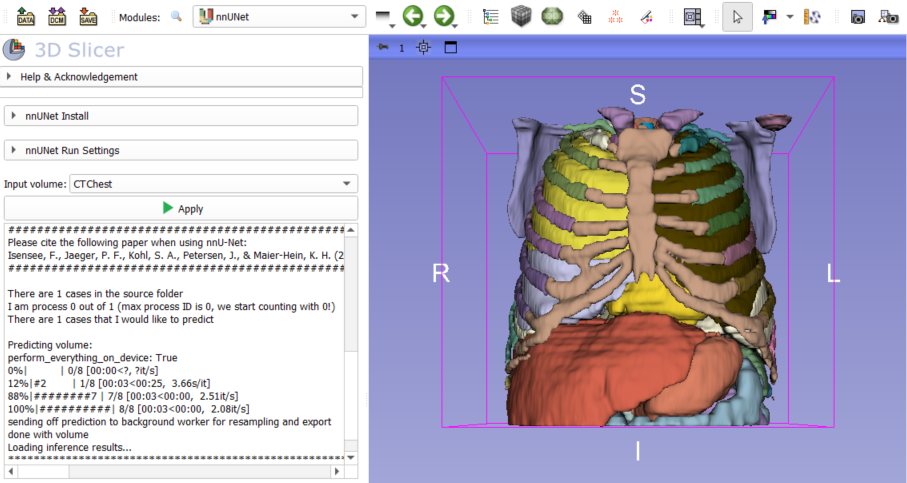
Select the volume on which to run the model using the volume input editor. Then click on the Apply button. The logs console will display all the information regarding running the input model.
Once the model has finished running, the segmentation will be loaded into 3D Slicer with its associated labels. It can then be viewed and edited in the Segment Editor module.
Creating custom nnU-Net extensions
To create your own nnU-Net extension, you can depend on and reuse this extension install logic and segmentation logic.
These components enable extension creators to quickly integrate nnUNet-based models in their own modules or custom 3D Slicer applications.
from SLicerNNUNetLib import InstallLogic
# Create instance
logic = InstallLogic()
# Connect progress logging
logic.progressInfo.connect(print)
# Get current version
logic.getInstalledNNUnetVersion()
# Install nnunetv2 requirements
logic.setupPythonRequirements()from SlicerNNUNetLib import SegmentationLogic, Parameter
# Create instance
logic = SegmentationLogic()
# Connect progress and error logging
logic.progressInfo.connect(print)
logic.errorOccurred.connect(slicer.util.errorDisplay)
# Connect processing done signal
logic.inferenceFinished.connect(logic.loadSegmentation)
# Start segmentation on given volume node on 5 folds
param = Parameter(modelPath=Path(r"C:\<PATH_TO>\NNUnetModel\Dataset123_456MRI"), folds = "0,1,2,3,4")
import SampleData
volumeNode = SampleData.downloadSample("MRHead")
logic.setParameter(param)
logic.startSegmentation(volumeNode)Credits
This extension was originally co-financed by the Fédération Française d’Orthodontie (FFO) as part of the Dental Segmentator developments and the Cure Overgrowth Syndromes (COSY) RHU Project (ANR-18-RHUS-0005).
The installation steps are based on the work done in the Slicer Total Segmentator extension.
This extension interfaces 3D Slicer with the nnUNet library.
Isensee, F., Jaeger, P. F., Kohl, S. A., Petersen, J., & Maier-Hein, K. H. (2021). nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods, 18(2), 203-211.