Embracing Change in Digital Pathology: The Improved Large Image Capabilities for HistomicsTK
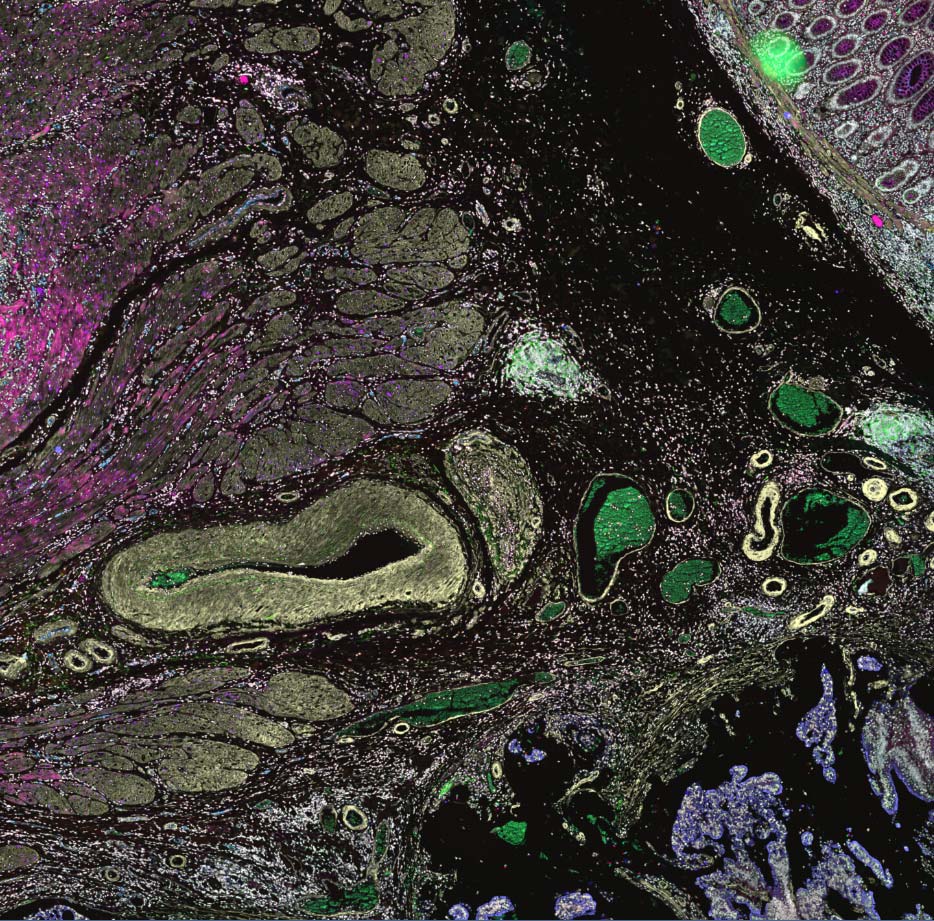
In the rapidly evolving field of digital pathology, Kitware’s open source software HistomicsTK stands out as a crucial platform for managing and disseminating extensive histopathology data.
Born from a collaboration with Emory University, HistomicsTK has evolved into a powerful, user-friendly web-based platform for digital pathology data. It played a vital role in accessing and interpreting the data from The Cancer Genome Atlas (TCGA), a comprehensive public resource encompassing a diverse range of cancers.
Built on the powerful open source data management platform Girder, HistomicsTK revolutionizes the world of digital pathology. By providing access to data via web APIs, user/group authentication, and fine-grained data access control, it ensures streamlined data handling, reliable security, and granular control over data visibility. These attributes contribute to more efficient operations and enhanced data protection, critical elements in an increasingly data-driven field.
Moreover, HistomicsTK brings innovative features specifically designed for digital pathology. Gallery views of whole-slide images and a multi-resolution image viewer facilitate deeper, more comprehensive image analyses, effectively enhancing pathologists’ diagnostic accuracy and capability. Custom metadata allows for better data organization and increased searchability, simplifying data management. A searchable interface delivers a seamless user experience, making relevant information easily accessible.
The Large Image module, an integral part of HistomicsTK, offers visualization of large, multi-resolution images overlaid with annotations. Compatible with numerous Whole Slide Imaging (WSI) file formats means that it can process outputs from multiple whole slides and digital pathology scanners. It also provides a Python API for managing these images, including tiled access, metadata extraction, and a range of annotations.
With that foundation in mind, it’s time to highlight the recent innovative strides HistomicsTK has made, especially in large image management.
Tile Sources: Our Tile Sources continue to improve, notably for multiple formats such as DICOM, TIFF, and Rasterio. DICOM whole slide images are gaining traction in the pathology community. Rasterio is mostly used for geospatial sources but allows reading some common formats on a wider variety of operating systems than our other tile sources. TIFF files come in a broad range of varieties. Although we already utilize libtiff, openslide, and have an ome tiff specific variant that employs libtiff, by using the tifffile module as an additional source for reading TIFFs, we can do a more effective job in reading certain multi-region tiff images. One key example is the Leica SCN format, which can store multiple tissue regions as separate images.
HistomicsTK now also features an innovative test tile source comprising elements such as bands, axes, and bit depth. This new addition bolsters our platform’s robustness, adding depth to our testing capabilities.
For ease in handling fluorescence microscopy images, we’ve introduced a feature to parse vendor information from QPTIFF and ImageJ files. This feature streamlines working with such image formats, making them more user-friendly and efficient, saving practitioners invaluable time and resources.
Kitware packages binary wheels for Linux to allow easy installation of all these tile sources, including the latest advances in the venerable openslide library.
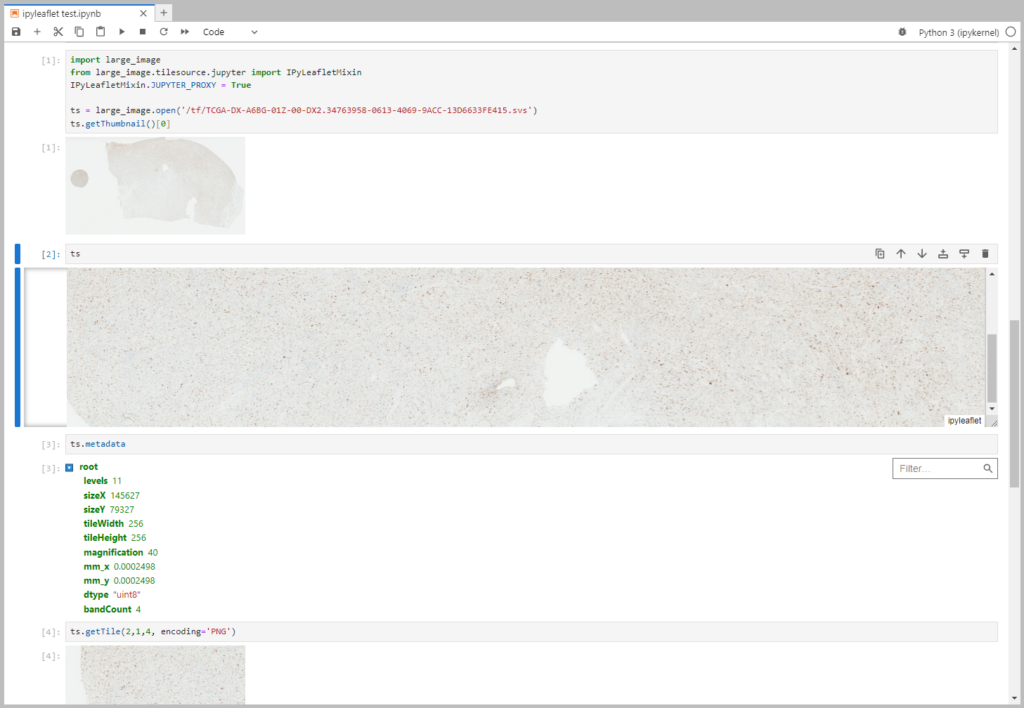
API: Enhancements include a canReadList function, improved class and output representation (python repr, see Figure 1), and properties like frame, dtype, and bandCount. In addition to these, the API has improved getPixel and histograms functions.
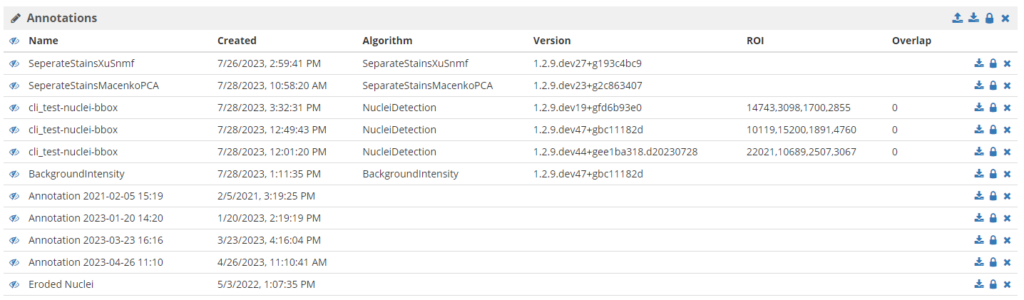
Features: HistomicsTK has made important strides in Girder data management and HistomicsUI. Metadata from items can be shown in sortable and filterable lists for efficient data navigation and searching as shown in Figure 2. Enhancements include lighter caching for improved system performance, inheritable config files for versatile management, and folder-level annotation access controls for enhanced security. Leveraging the power of inheritable config files, HistomicsTK now empowers each folder and collection to display distinct metadata and manage distinct annotation groups; these can even be changed per user or user group. Additionally, with the addition of new “style functions”, a Python function can be applied to tiles in real-time as they are served or read, offering a dynamic and responsive approach to data management.
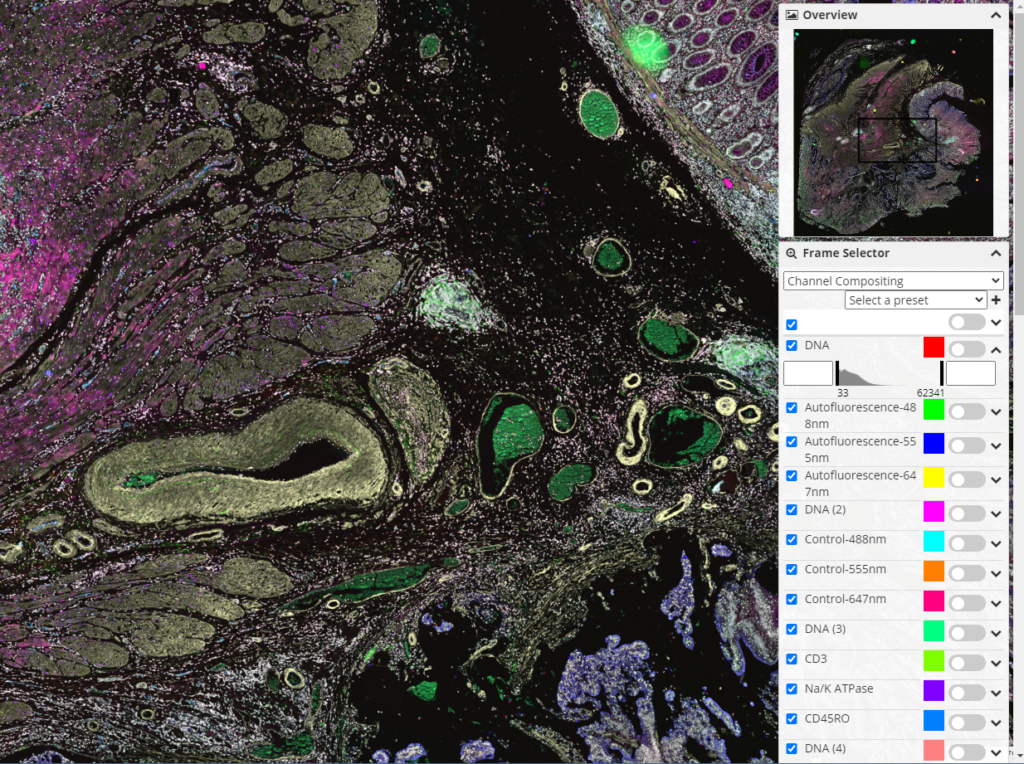
Other noteworthy additions include an inline YAML/JSON editor for straightforward data modification and metadata/annotation metadata search modes for ease of data discovery. HistomicsTK also supports ICC Color Profile for better color accuracy in images, which makes it feasible to have consistent results from algorithms and boasts a refined frame selector/histogram range (see Figure 3) for a more user-friendly experience. These updates significantly streamline operations in digital pathology.
Output Formats: HistomicsTK can now support all tile output formats that PIL can handle, including style range options.
Bug Fixes: Issues with iterating tiles with overlap, token scopes, and ETag use have been addressed.
Performance: HistomicsTK has a fallback for notification streams, improving the overall performance.
Together, these advancements make significant strides forward in digital pathology, offering users greater flexibility and efficiency in managing large volumes of histopathology data. The ongoing evolution of HistomicsTK epitomizes the relentless drive for innovation, striving to provide the most effective solutions to meet the changing needs of the histopathology community.
As the digital pathology industry continues to expand, HistomicsTK is committed to remaining at the forefront, constantly adapting and enhancing its capabilities to meet and exceed the demands of the field. To our valued commercial customers and partners, we look forward to embracing the future of histopathology together with HistomicsTK.
You can download HistomicsTK for free to access the new features of the Large Image module. For guidance and advanced support, you can reach out to our team of experts at Kitware. We can help customize the software according to your specific needs and project requirements, enabling you to leverage HistomicsTK effectively. Contact us for more information.
The research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number U24CA194362. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.