Harness the Power of Statistical Shape Models with Open Source and Kitware Custom Solutions
What is a Statistical Shape Model?
A statistical shape model (SSM), is a mathematical representation of the geometric variability of anatomical structures, such as bones or organs, in a population. This article presents an overview of SSMs and their importance for biomedical engineering and research, including deep learning and digital twin technologies. Both SSMs and digital twins are instrumental in the advancement of precision medicine, where combining detailed anatomical information with other patient-specific data allows healthcare providers to create highly personalized treatment plans. We also describe two open source software packages for computing SSMs, and how Kitware can help your company or research program integrate shape modeling into your applications.
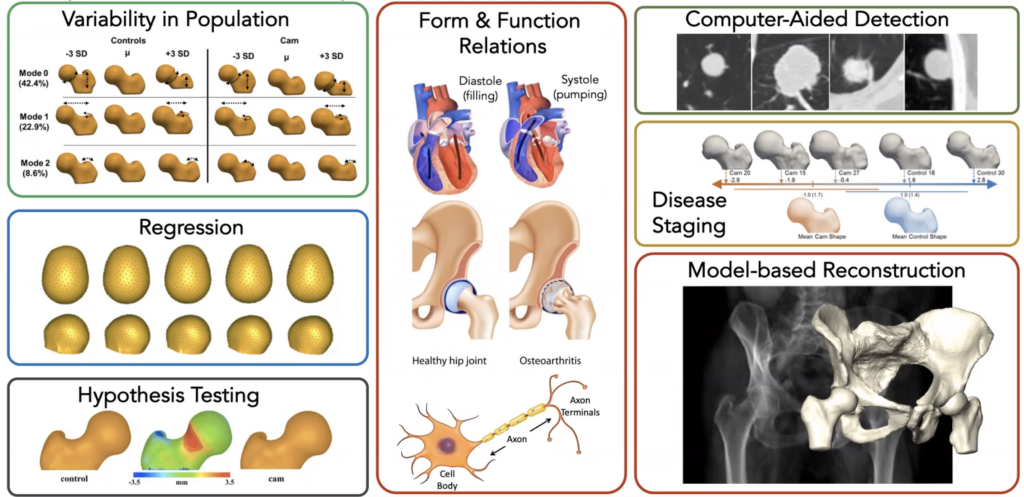
There are many important applications of SSMs to biomedical research and product development. In biomedicine, a SSM of patient hearts, bones, brain structures or other anatomy can help researchers and clinicians understand the range of normal anatomic shapes and sizes, and how those shapes vary with pathologies and genetic differences. Some examples include orthopedic implant design [1, 2], biomechanical simulation [3, 4], 3D reconstruction of patient anatomy [5 – 8], predictive analytics [9 – 11], and medical image processing [12 – 16]. SSMs are also being used to regularize and improve the quality of learning-based generative medical image models [17, 18]. More generally, SSMs are invaluable tools for basic scientific investigation, such as the role of specific genes in growth and patterning [19 – 21], characterizing normal and pathologic musculoskeletal phenotypes [22, 23], and measuring brain structure changes in dementia and other cognitive disorders [24 – 26].
SSMs are the fusion of traditional morphometrics with modern data-driven machine learning approaches. Perhaps the most well known early example of shape analysis is Darwin’s study relating the form and function of the varieties of Galapagos finch beaks, which helped lead to the theory of natural selection [27]. In the early 20th century, researchers formalized shape modeling into a field of mathematics known as morphometrics. Morphometrics, in turn, was influential in the development of modern multivariate statistical methods and is still in widespread use today. Modern SSMs employ a variety of algorithms to automatically quantify and measure anatomical shapes, including parametric and surface-sampling representations, which are optimized to produce population models with desirable properties for statistics. SSM advances offer automated approaches to modeling shape with a high degree of precision and detail.
SSMs and Digital Twins
SSMs are a fundamental component for building anatomical Digital Twins. Though distinct concepts, SSMs and Digital Twins are closely related and complementary in the context of medicine, particularly in the fields of radiology, orthopedics, and cardiology, where imaging plays an important part of clinical care. Typically derived from annotated and labeled medical image data, SSMs capture the variability in shapes across different individuals. Thus, they can be used as a rich source of statistical information for creating anatomical digital twins in medicine. SSMs can also be used to generate a specific instance that matches an individual patient’s anatomy, or to generate randomized simulated data. Such “generative models” are a crucial component of an anatomical digital twin. For example, in creating a digital twin of a patient’s heart, statistical shape modeling helps in understanding the range of normal and abnormal heart shapes and sizes, allowing the digital twin to accurately reflect the patient’s unique cardiac structure. This approach is also beneficial in complex surgical planning, prosthetic design, and targeted therapies in oncology.
Statistical shape modeling can also represent dynamic anatomy. In addition to static snapshots of anatomical shape, an SSM can also model the population statistics of anatomical shape over time, such as the change in the shape of the chambers of the heart over the cardiac cycle or a longitudinal model of shape changes over a period of growth or disease progression. Combining such “dynamic” shape models with physiological and biomechanical data creates a correspondingly “dynamic” Digital Twin for simulation and analysis of how patients might respond to different treatments or how anatomy changes over the course of a disease progression.
Open Source Tools for SSMs
ShapeWorks and SlicerSALT are open source software tools for creating SSM from your data. Kitware’s engineers are closely involved with two SSM software projects that are freely available for you to download and use. Both toolkits work directly from segmentations of image data and have an active community of developers and users for support.
ShapeWorks
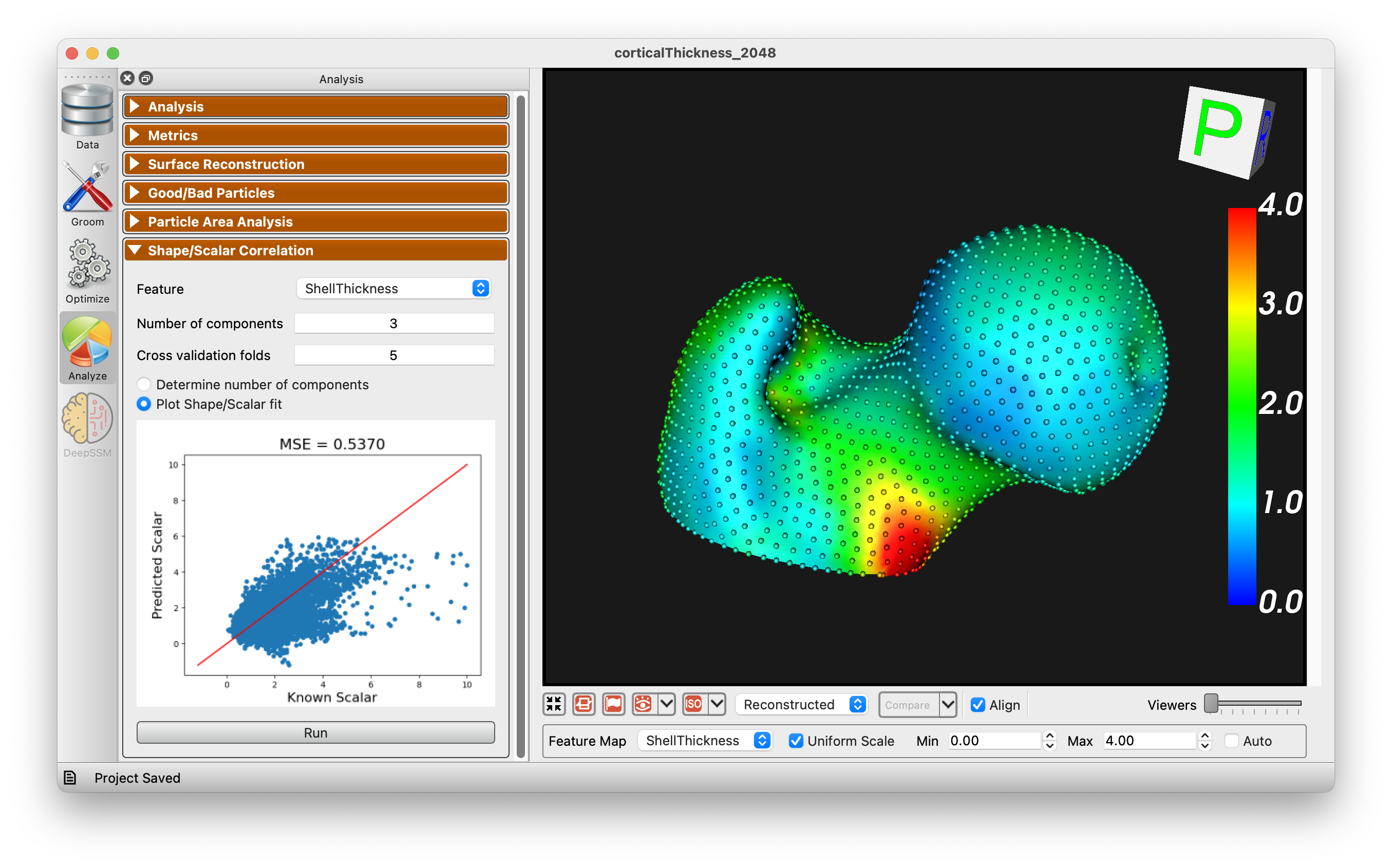
ShapeWorks [28] is a free, open-source suite of software tools that uses an automated surface sampling method for construction of landmark-based SSMs. It also includes software for model visualization and analysis. The ShapeWorks approach is particularly useful for applications that model the surface geometry of anatomy, including many orthopedic applications. Unique features of ShapeWorks allow it to handle very complex topologies, such as the pelvis. It can be used to construct longitudinal models and joint statistical models of anatomy that characterize the variability of one anatomical structure with respect to another, such as the bones comprising the knee or the four chambers of the heart. ShapeWorks is maintained and distributed by the University of Utah, with significant ongoing contributions from new research and collaborations and support from the NIH1.
SlicerSALT
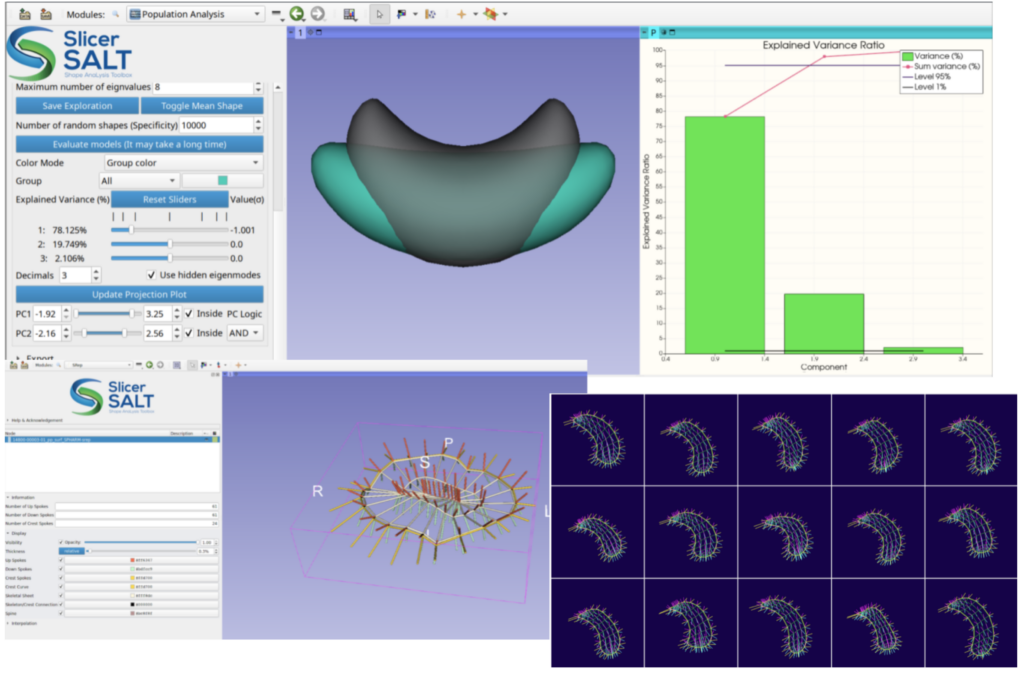
The Slicer Shape AnaLysis Toolbox (SlicerSALT) [29, 30] is a free, open source platform for developing and disseminating powerful SSM methodologies. It is distributed as a custom application built from 3D Slicer as well as a series of 3D Slicer Extensions. SlicerSALT includes support for spherical-harmonic and skeletal shape representations, non-Euclidean geometry statistics, and longitudinal shape modeling. It provides a user-friendly set of tools for visualization and analysis. Being based on 3D Slicer allows for easy integration with medical image segmentation, visualization, and analysis workflows. SlicerSALT also includes the newly developed SlicerPipelines functionality for creating reproducible and distributable workflows via 3D Slicer with no or minimal coding. SlicerSALT is developed and maintained by scientists and engineers at Kitware, with support from a community of collaborators2 and funding from the NIH3.
Want to learn more? Kitware can help.
Whatever your shape modeling and analysis needs, Kitware can help! Kitware engineers and scientists are world experts in SSMs and include some of the original inventors of the technology behind ShapeWorks and SlicerSALT. As with all data analytics, a good understanding of how to apply the right SSM approach to the right training data is important for obtaining the most powerful and interpretable results. Our scientists have a long history of research and industry collaboration in shape modeling and analysis. We are excited to apply our deep knowledge of SSMs to your problems!
Recent and Selected References
- Ingrid Dupraz, Arthur Bollinger, Julien Deckx, Ronja Alissa Schierjott, Michael Utz, and Marnic Jacobs. Using statistical shape models to optimize TKA implant design. Applied Sciences, 12(3):1020, 2022.
- Nina Kozic, Stefan Weber, Philippe Buchler, Christian Lutz, Nils Reimers, Miguel A Gonzalez Ballester, and Mauricio Reyes. Optimisation of orthopaedic implant design using statistical shape space analysis based on level sets. Medical image analysis, 14(3):265–275, 2010.
- Luke G Johnson, Sara Bortolussi-Courval, Anjuli Chehil, Emily K Schaeffer, Colleen Pawliuk, David R Wilson, and Kishore Mulpuri. Application of statistical shape modeling to the human hip joint: a scoping review. JBI Evidence Synthesis, 21(3):533, 2023.
- Amy L Lenz and Rich J Lisonbee. Biomechanical insights afforded by shape modeling in the foot and ankle. Foot and Ankle Clinics, 28(1):63–76, 2023.
- Amogh Patil, Krishan Kulkarni, Shuqiao Xie, Anthony MJ Bull, and Gareth G Jones. The accuracy of statistical shape models in predicting bone shape: a systematic review. The International Journal of Medical Robotics and Computer Assisted Surgery, page e2503, 2023.
- Hsuan-Yu Lu, Cheng-Chung Lin, Kao-Shang Shih, Tung-Wu Lu, Mei-Ying Kuo, Song-Ying Li, and Horng-Chaung Hsu. Integration of statistical shape modeling and alternating interpolation-based model tracking technique for measuring knee kinematics in vivo using clinical interleaved bi-plane fluoroscopy. PeerJ, 11:e15371, 2023.
- Abhirup Banerjee, Ernesto Zacur, Robin P Choudhury, and Vicente Grau. Automated 3d whole-heart mesh reconstruction from 2d cine MR slices using statistical shape model. In 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), pages 1702–1706. IEEE, 2022.
- Lars C Ebert, Dana Rahbani, Marcel Luthi, Michael J Thali, Angi M Christensen, and Barbara Fliss. Reconstruction of full femora from partial bone fragments for anthropological analyses using statistical shape modeling. Forensic Science International, 332:111196, 2022.
- Erik T Bieging, Alan Morris, Lowell Chang, Lilas Dagher, Nassir F Marrouche, and Joshua Cates. Statistical shape analysis of the left atrial appendage predicts stroke in atrial fibrillation. The international journal of cardiovascular imaging, 37(8):2521–2527, 2021.
- Charlene Mauger, Kathleen Gilbert, Aaron M Lee, Mihir M Sanghvi, Nay Aung, Kenneth Fung, Valentina Carapella, Stefan K Piechnik, Stefan Neubauer, Steffen E Petersen, et al. Right ventricular shape and function: cardiovascular magnetic resonance reference morphology and biventricular risk factor morphometrics in UK biobank. Journal of Cardiovascular Magnetic Resonance, 21:1–13, 2019.
- Shuman Jia, Hubert Nivet, Josquin Harrison, Xavier Pennec, Claudia Camaioni, Pierre Jais, Hubert Cochet, and Maxime Sermesant. Left atrial shape is independent predictor of arrhythmia recurrence after catheter ablation for atrial fibrillation: A shape statistics study. Heart Rhythm O2, 2(6):622–632, 2021.
- Simon Bohlender, Ilkay Oksuz, and Anirban Mukhopadhyay. A survey on shape-constraint deep learning for medical image segmentation. IEEE Reviews in Biomedical Engineering, 2021.
- Felix Ambellan, Alexander Tack, Moritz Ehlke, and Stefan Zachow. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the osteoarthritis initiative. Medical Image Analysis, 52:109–118, 2019.
- Chen Zhao, Joyce H Keyak, Jinshan Tang, Tadashi S Kaneko, Sundeep Khosla, Shreyasee Amin, Elizabeth J Atkinson, Lan-Juan Zhao, Michael J Serou, Chaoyang Zhang, et al. St-v-net: Incorporating shape prior into convolutional neural networks for proximal femur segmentation. Complex & Intelligent Systems, 9(3):2747–2758, 2023.
- Rosana El Jurdi, Caroline Petitjean, Paul Honeine, Veronika Cheplygina, and Fahed Abdallah. High-level prior-based loss functions for medical image segmentation: A survey. Computer Vision and Image Understanding, 210:103248, 2021.
- Tobias Heimann and Hans-Peter Meinzer. Statistical shape models for 3d medical image segmentation: a review. Medical image analysis, 13(4):543–563, 2009.
- Amirhossein Bayat, Danielle F Pace, Anjany Sekuboyina, Christian Payer, Darko Stern, Martin Urschler, Jan S Kirschke, and Bjoern H Menze. Anatomy-aware inference of the 3d standing spine posture from 2d radiographs. Tomography, 8(1):479–496, 2022.
- Mokshagna Sai Teja Karanam, Tushar Kataria, Krithika Iyer, and Shireen Y Elhabian. Adassm: Adversarial data augmentation in statistical shape models from images. In: International Workshop on Shape in Medical Imaging, pages 90–104. Springer, 2023.
- Joshua Cates, Lisa Nevell, Suresh I Prajapati, Laura D Nelon, Jerry Y Chang, Matthew E Randolph, Bernard Wood, Charles Keller, and Ross T Whitaker. Shape analysis of the basioccipital bone in pax7-deficient mice. Scientific Reports, 7(1):17955, 2017.
- Dean C Adams, F James Rohlf, and Dennis E Slice. Geometric morphometrics: ten years of progress following the ‘revolution’. Italian Journal of Zoology, 71(1):5–16, 2004.
- Christian Peter Klingenberg. Morphometrics and the role of the phenotype in studies of the evolution of developmental mechanisms. Gene, 287(1-2):3–10, 2002.
- Penny R Atkins, Praful Agrawal, Joseph D Mozingo, Keisuke Uemura, Kunihiko Tokunaga, Christopher L Peters, Shireen Y Elhabian, Ross T Whitaker, and Andrew E Anderson. Prediction of femoral head coverage from articulated statistical shape models of patients with developmental dysplasia of the hip. Journal of Orthopaedic Research, 40(9):2113–2126, 2022.
- Michael D Harris, Manasi Datar, Ross T Whitaker, Elizabeth R Jurrus, Christopher L Peters, and Andrew E Anderson. Statistical shape modeling of cam femoroacetabular impingement. Journal of Orthopaedic Research, 31(10):1620–1626, 2013.
- Sergi G Costafreda, Ivo D Dinov, Zhuowen Tu, Yonggang Shi, Cheng-Yi Liu, Iwona Kloszewska, Patrizia Mecocci, Hilkka Soininen, Magda Tsolaki, Bruno Vellas, et al. Automated hippocampal shape analysis predicts the onset of dementia in mild cognitive impairment. Neuroimage, 56(1):212–219, 2011.
- Kai-kai Shen, Jurgen Fripp, Fabrice Meriaudeau, Gael Chetelat, Olivier Salvado, Pierrick Bourgeat, Alzheimer’s Disease Neuroimaging Initiative, et al. Detecting global and local hippocampal shape changes in Alzheimer’s disease using statistical shape models. Neuroimage, 59(3):2155–2166, 2012.
- Raffaele Cacciaglia, Frauke Nees, Oliver Grimm, Stephanie Ridder, Sebastian T Pohlack, Slawomira J Diener, Claudia Liebscher, and Herta Flor. Trauma exposure relates to heightened stress, altered amygdala morphology and deficient extinction learning: Implications for psychopathology. Psychoneuroendocrinology, 76:19–28, 2017.
- Charles Darwin. On the origin of species, 1859, 2016.
- Joshua Cates, Shireen Elhabian, and Ross Whitaker. Shapeworks: particle-based shape correspondence and visualization software. In Statistical Shape and Deformation Analysis, pages 257–298. Elsevier, 2017.
- Jared Vicory, Ye Han, Juan Carlos Prieto, David Allemang, Mathieu Leclercq, Connor Bowley, Harald Scheirich, Jean-Christophe Fillion-Robin, Steve Pizer, James Fishbaugh, et al. Slicersalt: From medical images to quantitative insights of anatomy. In International Workshop on Shape in Medical Imaging, pages 201–210. Springer, 2023.
- Jared Vicory, Laura Pascal, Pablo Hernandez, James Fishbaugh, Juan Prieto, Mahmoud Mostapha, Chao Huang, Hina Shah, Junpyo Hong, Zhiyuan Liu, et al. Slicersalt: Shape analysis toolbox. In Shape in Medical Imaging: International Workshop, ShapeMI 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings, pages 65–72. Springer, 2018.
- ShapeWorks is supported by the National Institutes of Health under grant numbers NIBIB-U24EB029011, NIAMS-R01AR076120, NHLBI-R01HL135568, NIBIB-R01EB016701, and NIGMS-P41GM103545.
↩︎ - https://salt.slicer.org/citations/ ↩︎
- SlicerSALT is supported by the National Institutes of Health under grant number R01EB021391
↩︎