Introducing SlicerPET 2.0
As noted in the paper “SlicerPET: A workflow based software module for PET/CT guided needle biopsy” [1], clinicians frequently use biopsies to confirm cancer diagnoses when radiologically indicated. Positron emission tomography/computed tomography (PET/CT) guidance may improve the diagnostic yield of biopsies, as PET can localize tumors that are not visible in CT. To bring PET image guidance into the interventional CT suite [2], Kitware developed a user-friendly, workflow-based application, known as SlicerPET.
In this article, we summarize the work detailed in the paper and introduce a new version of the software application (SlicerPET 2.0).
Workflow Steps
SlicerPET utilizes a module [1] to facilitate PET/CT-guided needle biopsy. The module is implemented using the 3D Slicer framework [3]. 3D Slicer is a software platform for medical image analysis and visualization. Medical researchers employ 3D Slicer to visualize and process images.
Following the development of 3D Slicer 4.5 [4], Kitware released SlicerPET 2.0. SlicerPET 2.0 takes advantage of the improvements and bug fixes made in 3D Slicer 4.5.
SlicerPET 2.0 introduces an all-new logo and icon.
Both 3D Slicer and SlicerPET’s module are open source. The following subsections outline the workflow of the module.
The workflow begins with data loading.
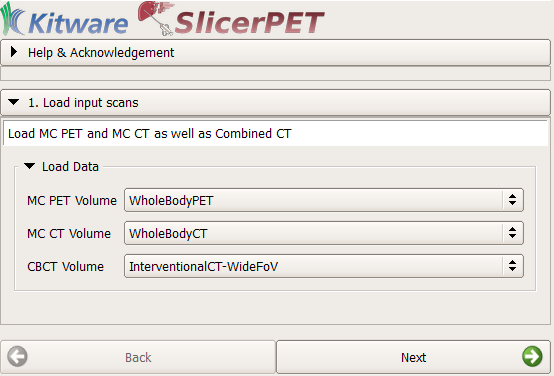
Step 1: Data Loading
The workflow begins with the selection of three volumes required for needle guidance. These volumes are respiratory-compensated PET, respiratory-compensated CT, and interventional CT scans. Any of the image-loading techniques available in 3D Slicer work when making this selection. Examples include use of the Digital Imaging and Communications in Medicine (DICOM) database browser, as well as drag and drop.
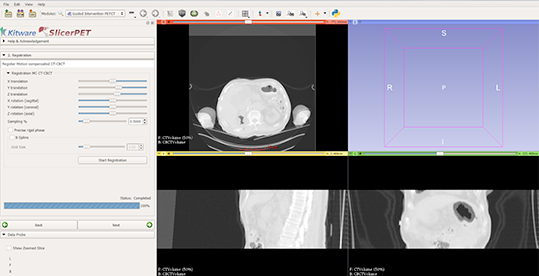
Step 2: Registration
SlicerPET registers the respiratory-compensated CT with the interventional CT scan. SlicerPET 2.0 features a refactored registration step, which allows for multi-phase rigid body registration in addition to optional deformable B-spline registration. The release also offers a manual interface for adjusting the initial transform to ensure convergence, even with grossly different CT images. What is more, the release removes rarely used parameters from the user interface.
Completing rigid-only registration makes adjusted initial positions visible in the left panel. The slice views show an overlap of interventional CT and CT components from the PET/CT suite.
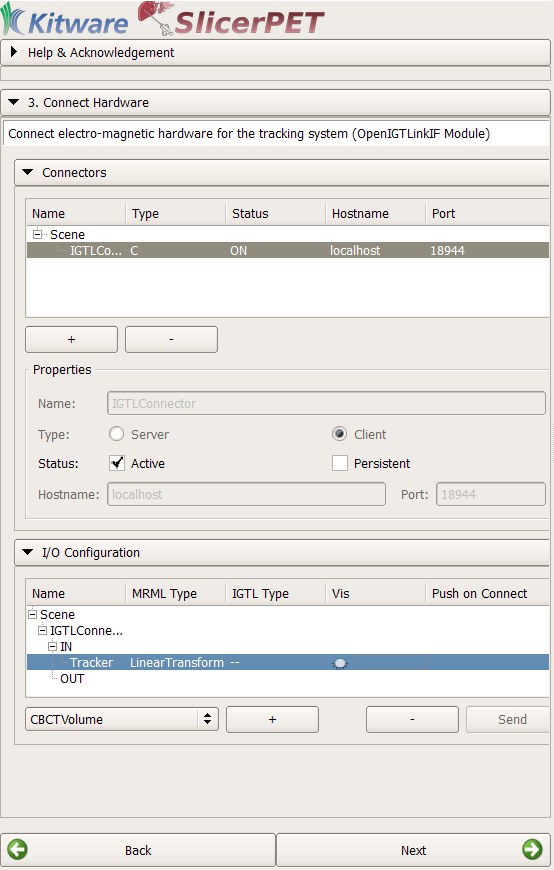
Step 3: Tracking
SlicerPET establishes a connection between the tracking system and 3D Slicer using the Image Guided Surgery Toolkit (IGSTK). IGSTK is a component-based framework that reads and displays medical images. The toolkit offers visualization and point-based registration capabilities, with provisions for logging, handling exceptions, and completing problem resolution.
SlicerPET uses IGSTK to connect the tracking device with 3D Slicer.
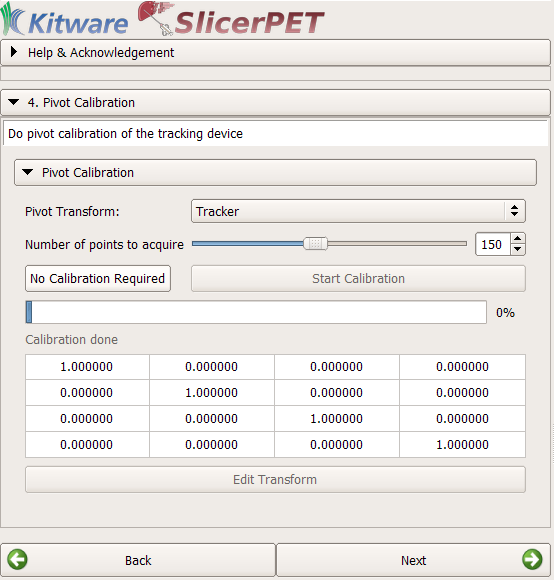
Step 4: Tool Calibration
If necessary, SlicerPET finds the transformation between the tip and the head of the biopsy needle. To do so, SlicerPET relies on Public software Library for UltraSound imaging research’s (PLUS’s) PivotCalibration module [5].
The pivot-calibration step is optional.
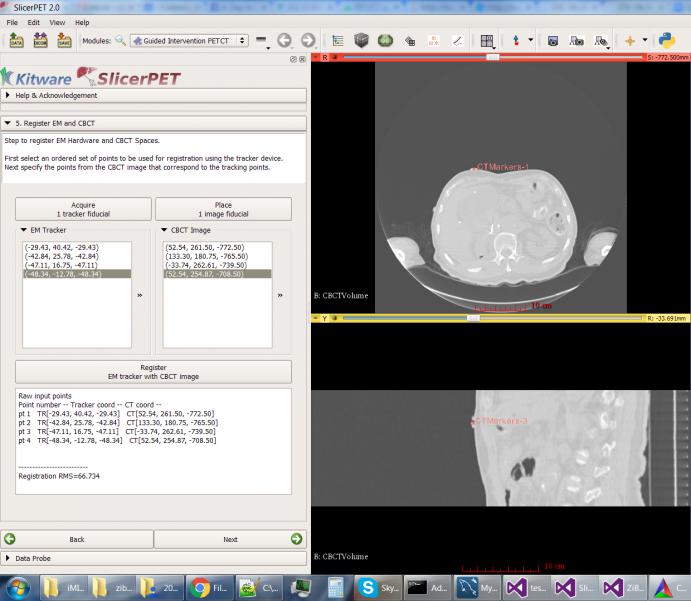
Step 5: Patient Registration
Through point-based registration, SlicerPET registers the interventional CT coordinate system to the needle tracker coordinate system in the interventional suite.
The test session captured in the screenshot uses a simulated tracker, which moves randomly. The high root mean square (RMS) error reflects this movement.
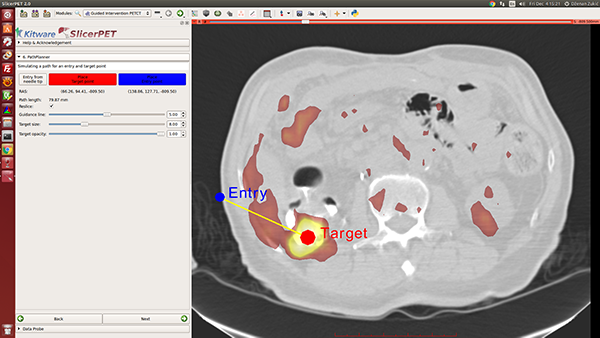
Step 6: Needle Path Planning
Once given target and entry points, SlicerPET visualizes the path between them. Repositioning the endpoints enables avoidance of ribs, etc., during procedures. A key highlight of this step is the PET image layer, which makes it easy to localize tumors.
SlicerPET permits adjustment of planned-path visualization parameters.
SlicerPET 2.0 improves visualization elements in needle path planning. The next step, guidance, also benefits from improved visual elements.
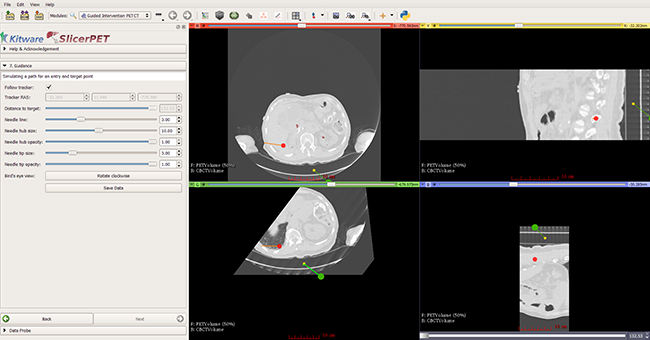
Step 7: Guidance
In this step, SlicerPET overlays a model of the needle and offers 3D visualization guidance for needle placement.
During a biopsy procedure, SlicerPET tracks the needle in real time and updates the virtual image. SlicerPET displays four slices: axial, sagittal, parallel to the needle, and perpendicular to the needle. Capabilities of the latter display include rotation. Clinicians use this display as guidance for biopsies.
SlicerPET allows for adjustment of needle visualization parameters.
With version 2.0, SlicerPET can save statistical data and a screenshot at the end of each procedure. Clinicians refer to this information for quantitative analysis of biopsy guidance.
A clinical feasibility study at Georgetown University is underway to evaluate the effectiveness of the SlicerPET guidance system for clinical patients. For more information on SlicerPET, contact kitware@kitware.com. To use the latest version of SlicerPET, download the installers [6] and the source code [7].
Acknowledgment
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R42CA153488. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
1. Zukić, Dženan, Julien Finet, Emmanuel Wilson, Filip Banovac, Giuseppe Esposito, Kevin Cleary, and Andinet Enquobahrie. “SlicerPET: A workflow based software module for PET/CT guided needle biopsy.” Proceedings of the Computer Assisted Radiology and Surgery 29th International Congress and Exhibition, Barcelona, Spain, June 24-27, 2015: S186-S187.
2. Kitware, Inc. “Overview.” PET-CT. http://public.kitware.com/Wiki/PET-CT.
3. Fedorov Andriy, Reinhard Beichel, Jayashree Kalpathy-Cramer, Julien Finet, Jean-Christophe Fillion-Robin, Sonia Pujol, Christian Bauer, Dominique Jennings, Fiona Fennessy, Milan Sonka, John Buatti, Stephen Aylward, James V. Miller, Steve Pieper, and Ron Kikinis. “3D Slicer as an Image Computing Platform for the Quantitative Imaging Network.” Magnetic Resonance Imaging 30 (2012): 1323-41. PMID: 22770690.
4. “Summary.” Documentation/4.5/Announcements. http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.5/Announcements.
5. Lasso, Andras. “Home.” Plus. November 25, 2015. https://www.assembla.com/spaces/plus/wiki.
6. Kitware, Inc. “SlicerPET.” Girder. http://bit.ly/1Inw5LF.
7. “SlicerPET.” Petct / SlicerPET. https://gitlab.kitware.com/petct/SlicerPET.
Dženan Zukić is a research and development engineer on Kitware’s Medical Computing team. His interests include image processing and manipulation, as well as image-guided surgical software.
Emmanuel Wilson is a staff scientist at Children’s National Health System in Washington, DC. His interest is in biomedical technology development for the clinical environment.
Kevin Cleary leads an interdisciplinary bioengineering team at Children’s National Health System, with a focus on improving visualization in pediatric surgery through medical devices and robotics.
Andinet Enquobahrie is the Assistant Director of Medical Computing at Kitware. As a subject matter expert, he is responsible for technical contribution and management of image-guided intervention, quantitative imaging, and surgical simulation projects.