SlicerSALT: ShapeVariationAnalyzer for Skeletal Representations
We have added new functionality to the ShapeVariationAnalyzer (SVA) module under SlicerSALT for accepting skeletal representations (s-reps). Following our previous blog post on enabling ShapePopulationViewer (SPV) to accept s-reps, users can now import multiple groups of s-reps and perform principal component analysis (PCA) in SVA. The process of performing PCA under SVA is covered below.
Creation of CSV File for Classification Groups
The PCA process in SVA starts with the creation of a CSV file containing the list of input geometries and their group information (Figure 1). SVA requires each group of input geometries to be located under the same folder. For s-reps, the header files (with extension “.xml”) are required to be in the same folder with absolute or relative paths for up/down/crest spokes correctly specified in those headers. The folders should not contain any other files ending with “.vtk”, which otherwise will be added to the CSV file if presented. The spoke geometry files are stored with extension “.vtp”, so it is fine to put them under the same folder as their headers. After adding groups of s-reps, click on “Export” to create a CSV file, which records the paths to the s-rep headers and their group information.
Figure 1: Load folders containing s-reps into SVA.
Preview/Update Groups
After the CSV file is created, SVA will automatically load it into a table. Users can then change and update the group assignment for the s-reps in SVA if needed (Figure 2).
Figure 2: Change group assignment and select s-reps for previewing.
After users click on the “Preview” button, SPV will pop up and show the header files toggled under the “Visualization” column (Figure 3).
Figure 3: Previewing s-reps of hippocampi under different groups in SPV.
Similar to the regular geometries, temporary vtkPolyData files will be created under Slicer’s temporary folder for previewing s-reps across groups. For better visualization purposes, the cellType attribute is also attached for identifying spoke types in SPV (Figure 4).
Figure 4: Use the cellType attribute to identify spoke types of the s-reps.
PCA
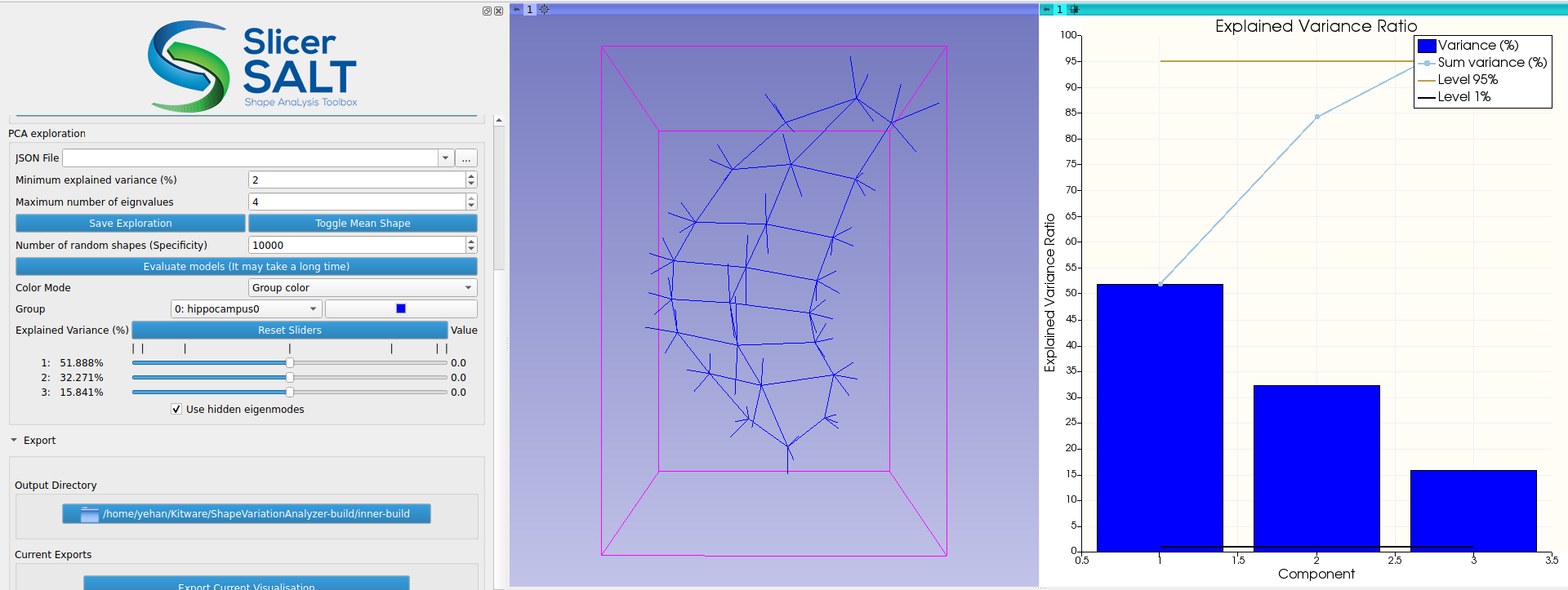
For regular geometries, PCA is performed directly on the point positions in SVA. Because s-reps are non-Euclidean, the s-rep data are first transformed into a new space called Z-space using composite principal nested spheres (CPNS). Then PCA is performed on the CPNS-transformed s-rep data in Z-space. As demonstrated in the work by Pizer et al., applying CPNS on s-reps could yield a more compact representation, meaning that fewer components are needed for capturing the majority of the variations of the input s-reps. After PCA is performed, users can interactively visualize how the weights of the principal components change the s-rep and compare the deformed s-rep with mean shape using the “Toggle Mean Shape” button (Figure 5). The PCA results can be saved using the “Save Exploration” button and loaded using the “JSON File” entry.
Figure 5: Visualizing the change in s-rep interactively by varying the weights of the principal components under Z-space.
Future Work
- We will add functionalities for computing compactness/specificity/generalization for model evaluation under SVA.
- The current loading process requires re-computing the CPNS model based on the inputs due to the complexity in storing the multi-component CPNS model. We will work on a compatible way of saving the CPNS model to the .json file to make the loading process more efficient.
- The current SVA does not contain functionality for projecting unseen data onto current principal components. We plan to add this functionality for both regular geometries and s-reps.
What is SlicerSALT
SlicerSALT is comprehensive software that will allow biomedical scientists to precisely locate shape changes in their imaging studies. As an offspring of 3D Slicer, SlicerSALT is free to download and its code is open source. SlicerSALT can be used to create 3D models from the landmarks and display them in various views. The software’s interface does not require a technical background to navigate, so researchers outside of computer science can use it for morphometry analysis. This software will enhance the intuitiveness and ease of use for such studies, as well as allow researchers to find shape changes with higher statistical power. Potential fields of interest include pharmacology, neuroscience, paleontology, and orthodontics. Altogether this constitutes a crucial resource for the imaging field that will enable many important new findings in biomedical imaging studies.
More information about SlicerSALT is available at http://salt.slicer.org/.
Acknowledgements
In addition to Drs. Paniagua, Han and Vicory at Kitware, the s-rep work is based on the seminal work by Dr. Steve Pizer and others, at the Medical Image Display and Analysis Group (MIDAG).
The SlicerSALT developers gratefully acknowledge funding for the SlicerSALT application provided by NIH NIBIB R01EB021391 and R56EB021391 (PI. Paniagua, Shape Analysis Toolbox for Medical Image Computing Projects and Shape Analysis Toolbox: From medical images to quantitative insights of anatomy), as well as R01HL153166 (PI. Jolley Computer Modeling of the Tricuspid Valve in Hypoplastic Left Heart Syndrome).
The SlicerSALT suite was developed by Kitware’s Medical Computing Team. If you would like to learn more about the capabilities of our Medical Computing Team, please email us at kitware@kitware.com or visit our website.