Introducing the 3D Slicer PolycysticKidneySeg Extension
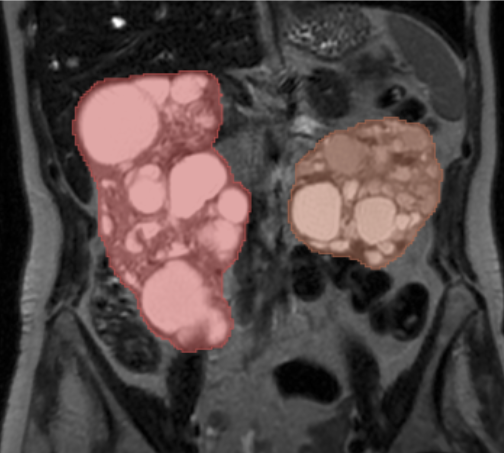
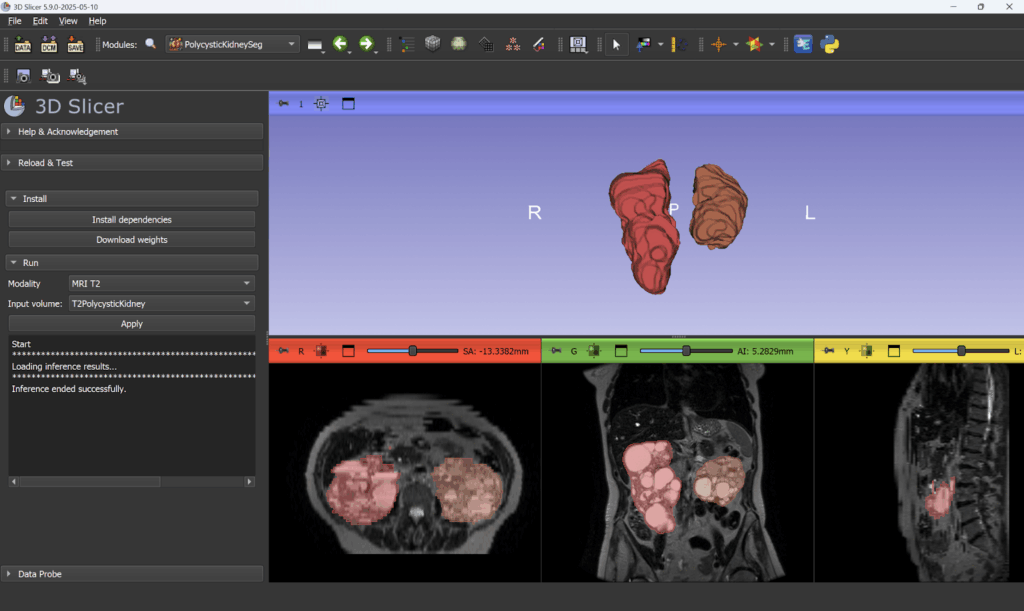
Kitware is pleased to announce the release of the PolycysticKidneySeg extension for 3D Slicer, which integrates the dual-task AI segmentation model for polycystic kidneys developed by Conze et al [1]. This extension enables researchers and clinicians to segment kidneys affected by Autosomal-Dominant Polycystic Kidney Disease (ADPKD) in both CT and T2-weighted MRI scans.
Understanding ADPKD and the Need for Advanced Segmentation
ADPKD is a common genetic disorder characterized by the development of numerous cysts in the kidneys, often leading to kidney enlargement and renal failure. Accurate and consistent kidney segmentation is critical for understanding disease progression, supporting clinical research, and improving patient monitoring.
Powered by State-of-the-Art AI
The segmentation model integrated into this extension is based on the work of Conze et al [1], who conducted a comprehensive evaluation of:
- Convolutional-based architectures
- Transformer-based architectures
- Hybrid convolutional/Transformer networks
They explored three training strategies:
- A single network to segment both kidneys
- Separate networks for each kidney
- A dual-task model with one shared encoder and two kidney-specific decoders
The best performance was achieved using the SwinUNetV2 architecture trained in a dual-task configuration—this is the model included in the extension.
Using the PolycysticKidneySeg Extension
The extension is compatible with 3D Slicer 5.9 preview release and later, and can be installed via the Extension Manager. Here’s how to get started:
- Install the extension from the 3D Slicer Extension Manager.
- Use the dedicated button to install required dependencies.
- Restart Slicer to finalize installation.
- On first use, download the pre-trained neural network weights.
- Select an input volume, specify the imaging modality (CT or T2), and press Apply to run the segmentation.
For practice and demonstration purposes, sample datasets are available in the Sample Data module:
- T2PolycysticKidney
- CTPolycysticKidney
You can explore the source code for the extension module on GitHub [2].
Acknowledgements
This extension was developed as part of research funded by the Société Francophone de Néphrologie, Dialyse et Transplantation (SFNDT). The underlying models were trained by Pierre-Henri Conze (IMT Atlantique) using data from the Genkyst cohort.
Kitware’s Role: Helping You Go Further, Faster
While 3D Slicer offers impressive capabilities out of the box, many research teams and product developers turn to Kitware for advanced customization and support. Our experts can help:
- Develop tailored extensions for your specific research or product needs.
- Integrate new algorithms and machine learning models.
- Support regulatory pathways with validated pipelines.
- Optimize performance for large datasets or real-time applications.
Explore our success stories:
- ✨ Customer Highlights: See how companies and industry partners are building commercial products with 3D Slicer.
- 🔬 Research and Development: Explore case studies that demonstrate how Slicer facilitates cutting-edge medical imaging research and product development.
We also maintain strong ties to the Slicer community and help guide the platform’s evolution through contributions, governance, and support services.
Contact our team to explore how we can help you achieve your goals.
References
[1] Conze PH, Andrade-Miranda G, Le Meur Y, Cornec-Le Gall E, Rousseau F. Dual-task kidney MR segmentation with transformers in autosomal-dominant polycystic kidney disease. Comput Med Imaging Graph. 2024 Apr;113:102349. DOI: 10.1016/j.compmedimag.2024.102349[2] PolycysticKidneySeg module accessed 05/17/2025
[2] github.com/conze/SlicerPolycysticKidneySeg