Accelerating the computation of shape representations in SPHARM-PDM

SPHARM-PDM is a module in SlicerSALT responsible for generating corresponding point-based shape models from sets of binary anatomical regions of interest (ROIs). It forms the foundation for much of SlicerSALT’s subsequent analysis and is one of our most widely used tools.
Originally, the concept behind SPHARM-PDM focused on establishing a one-to-one mapping between a 3D object’s surface and the surface of a unit sphere. This enabled explicit parametric representations of simply connected 3D surfaces. The code supporting this parameterization was later released as a set of command-line interface (CLI) modules. Since their registration with the NIH Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC) in 2009, 17 versions of the toolbox—ranging from v1.8 to v1.13—were released and downloaded 2,813 times. Since becoming a 3D Slicer extension in 2013, this tool has been downloaded over 100,000 times. SPHARM-PDM and SlicerSALT have been cited and mentioned in scientific literature thousands of times, underscoring their importance across numerous and diverse shape analysis projects in medical imaging.
Recently, we undertook a major overhaul of SPHARM-PDM’s C++ codebase, driven by the need for improved scalability, maintainability, and runtime performance. As part of this effort, we explored numerical precision, memory allocation strategies, and the limits of common Python and scientific computing frameworks to modernize the tool’s performance.
Our initial goal was to port the entire C++ codebase to Python. We created a prototype using NumPy and SciPy; however, scalability issues quickly surfaced. SPHARM-PDM’s parametric mapping requires solving large systems of equations, which—although structurally sparse—were represented as dense matrices. Operations on these dense matrices were prohibitively memory-intensive, and none of the sparse matrix options we tried (e.g., scipy.sparse, PyTorch autodiff, IPOPT, or Numba) yielded satisfactory performance improvements.
We pivoted back to optimizing and modernizing the existing C++ codebase. Adopting the Eigen library for sparse matrix operations and linear solvers delivered two key benefits: moderate but consistent runtime improvements and significantly cleaner, more maintainable code—with fewer raw pointers and simpler debugging. Additionally, we discovered that a surprising amount of runtime was being spent on memory allocation. By reusing memory and reducing redundant allocations—facilitated by Eigen—we further reduced runtime and streamlined control flow.

Precision tuning also played a critical role. We found that some linear systems were being solved with unnecessarily high precision. Striking the right balance was crucial: too little precision led to convergence issues, while excessive precision caused slowdowns. Tuning this appropriately led to substantial speedups.
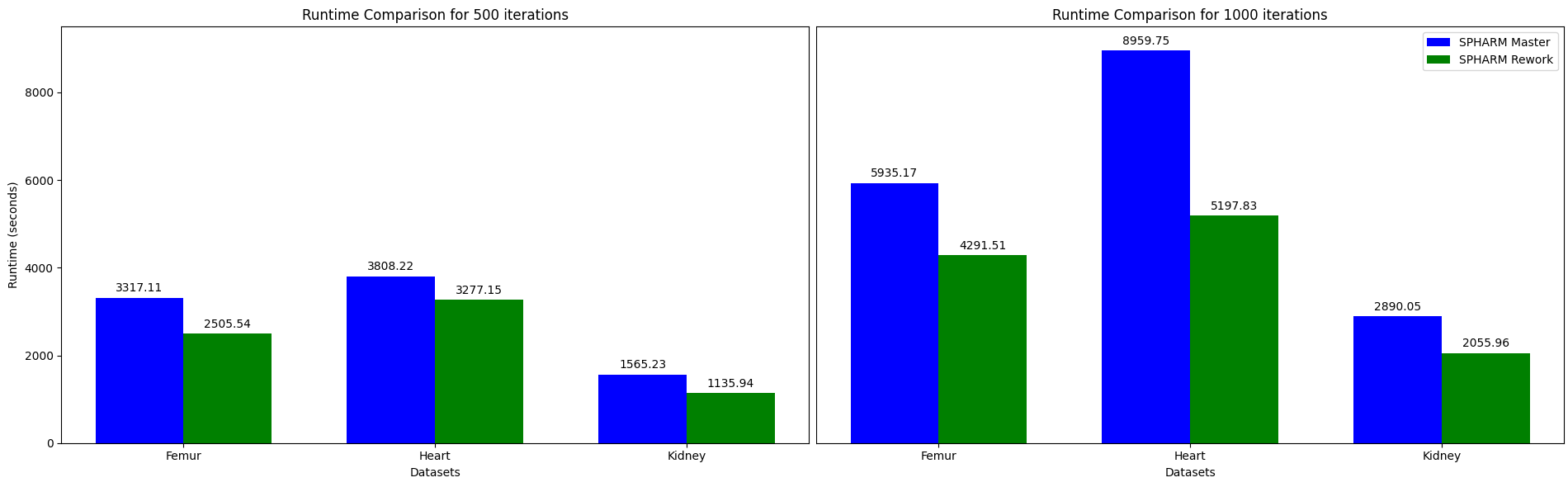
This rework offered valuable insights into the hidden costs of abstraction and the importance of selecting the right tools for the scale of the problem. Using Eigen significantly reduced raw pointer use, simplified memory management, and made the overall program flow more predictable and maintainable. These improvements collectively resulted in better convergence and in 30% to 40% speedups on real healthcare datasets of different sizes.
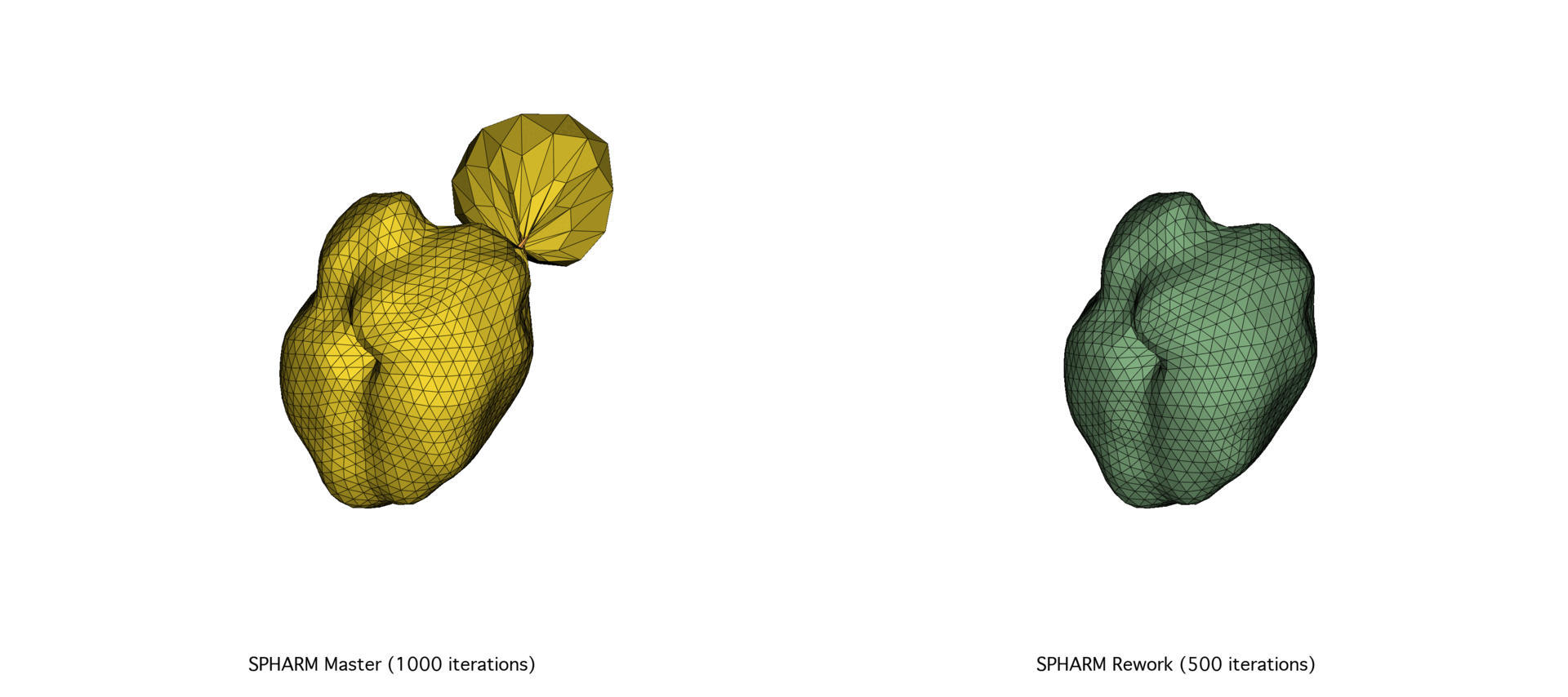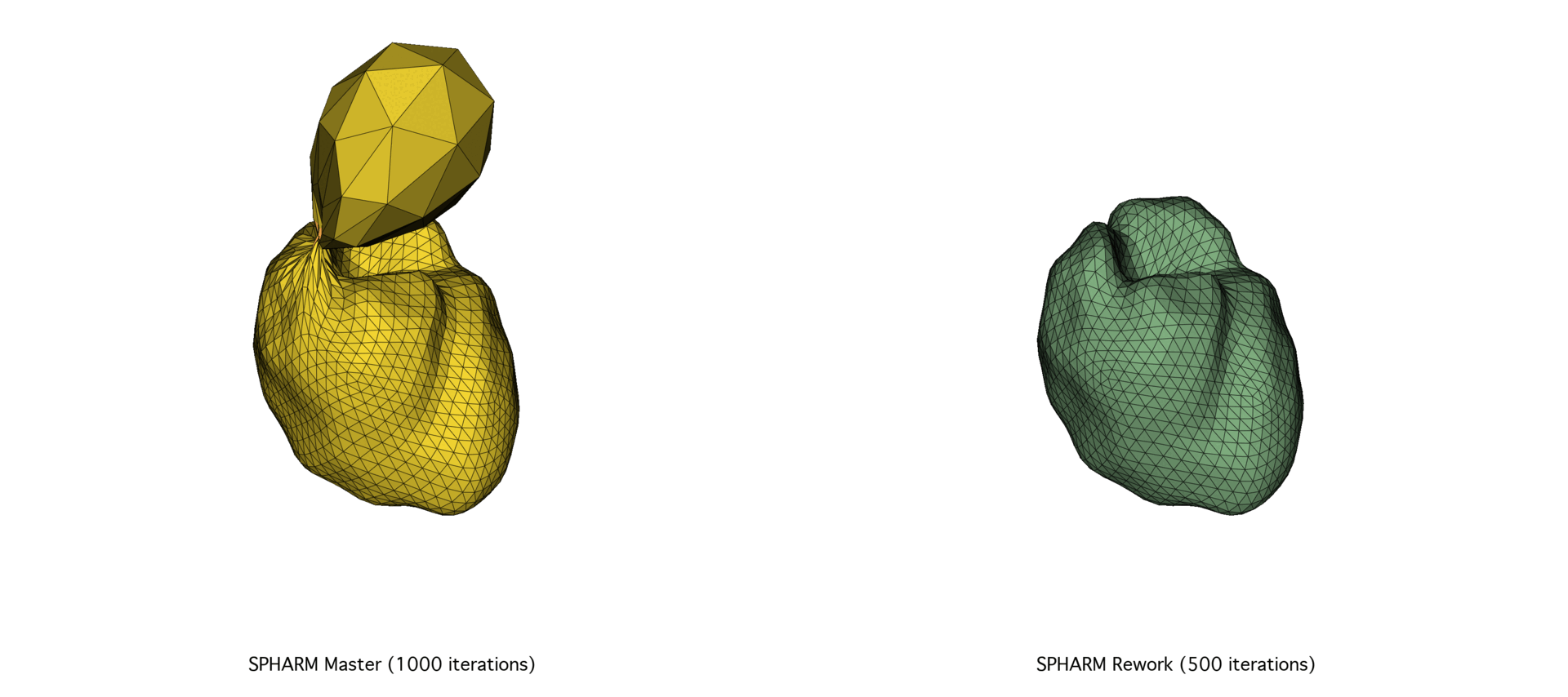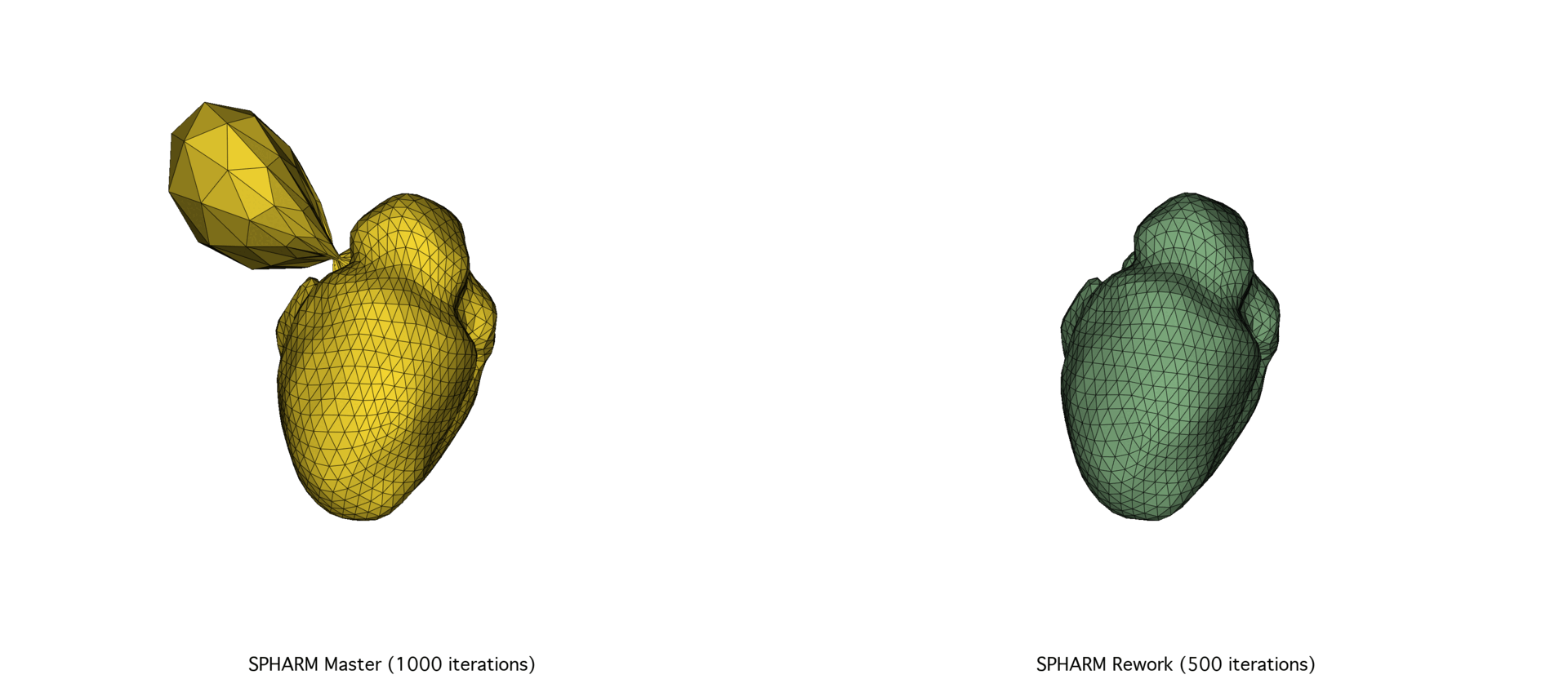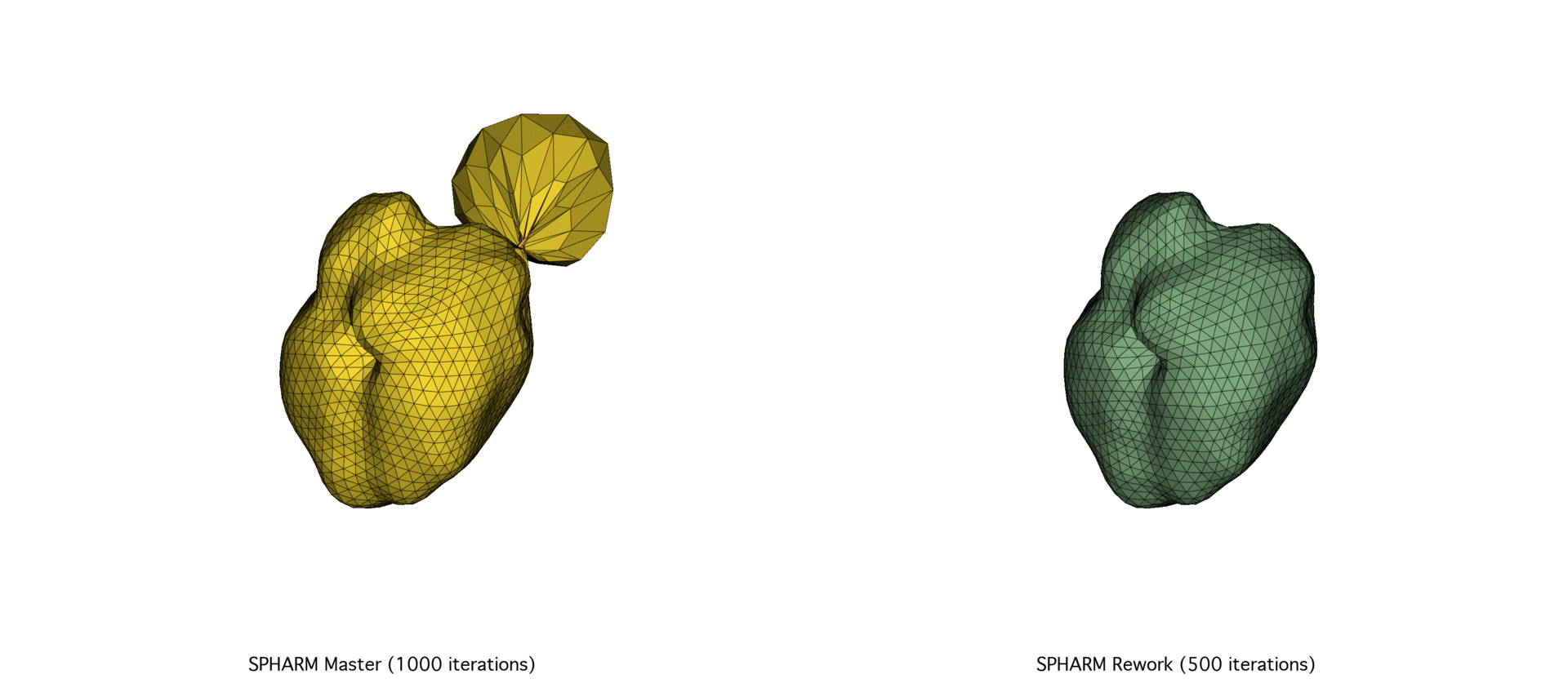
The continued evolution of SPHARM-PDM highlights the importance of pairing robust theoretical foundations with modern, high-performance computing practices. By reevaluating this tool’s implementation from the ground up, we not only improved its efficiency and maintainability but also ensured its long-term viability as a core component of SlicerSALT.
Acknowledgements
SlicerSALT is the product of a multidisciplinary collaboration between Kitware, the NYU Tandon School of Engineering, and the University of North Carolina at Chapel Hill. Its development is supported by the NIH National Institute of Biomedical Imaging and Bioengineering under grant R01EB021391 (Shape Analysis Toolbox for Medical Image Computing Projects).
Get in touch!
SlicerSALT exemplifies the type of custom 3D Slicer applications that Kitware develops. Contact us at kitware@kitware.com to learn how we can help integrate 3D Slicer-based solutions into your research, workflows, and products.