SlicerSALT 6.0 released

The SlicerSALT team is pleased to announce the latest release of SlicerSALT, which includes a comprehensive set of updates and advanced features aimed at enhancing the capabilities of shape analysis research. Version 6.0, now available for download, introduces exciting new functionality detailed below.
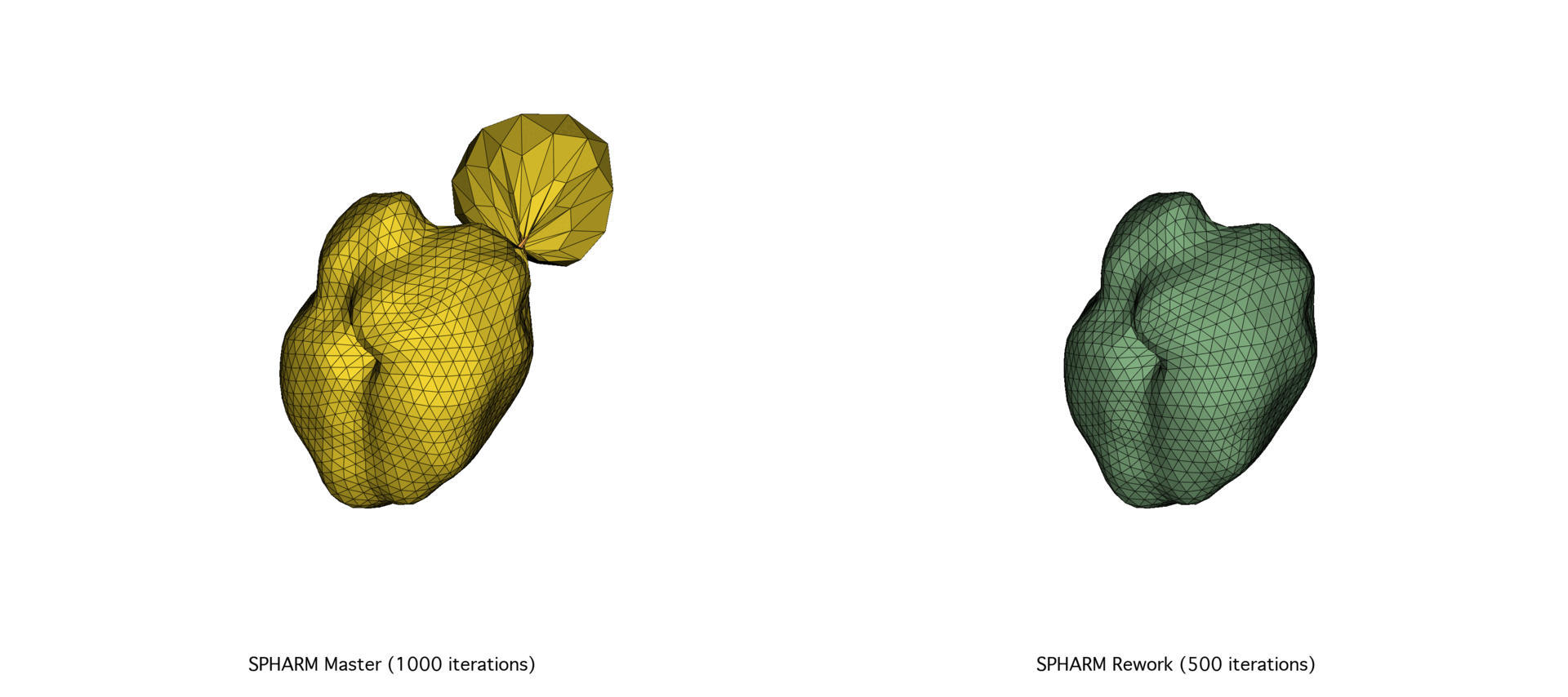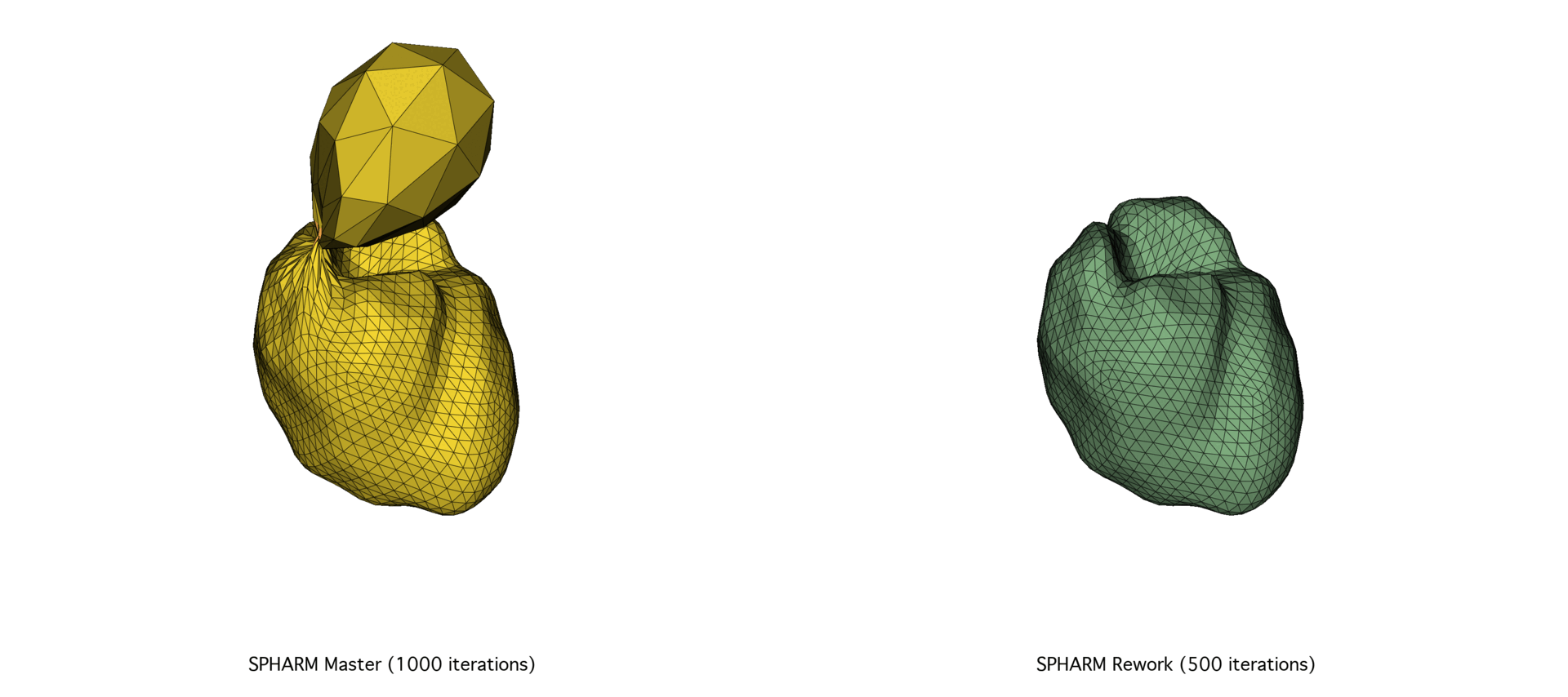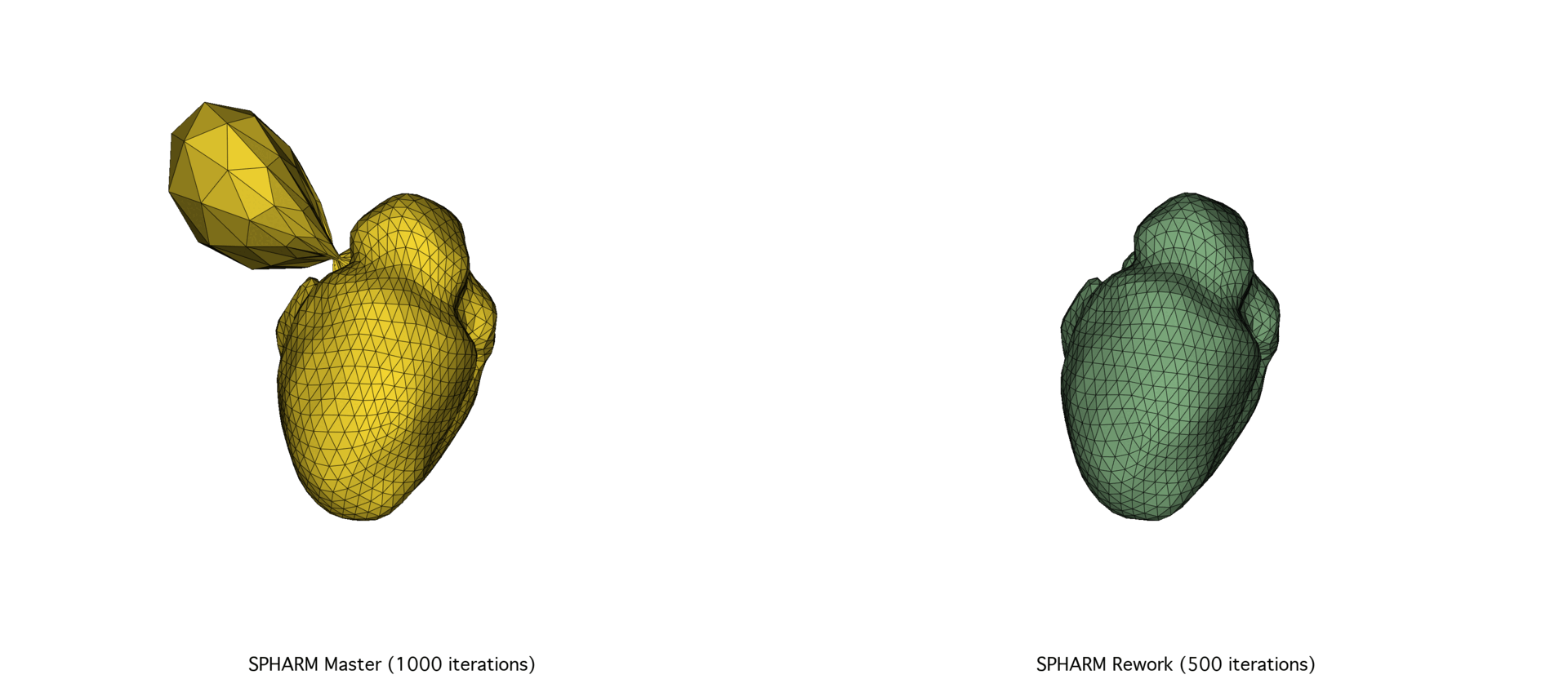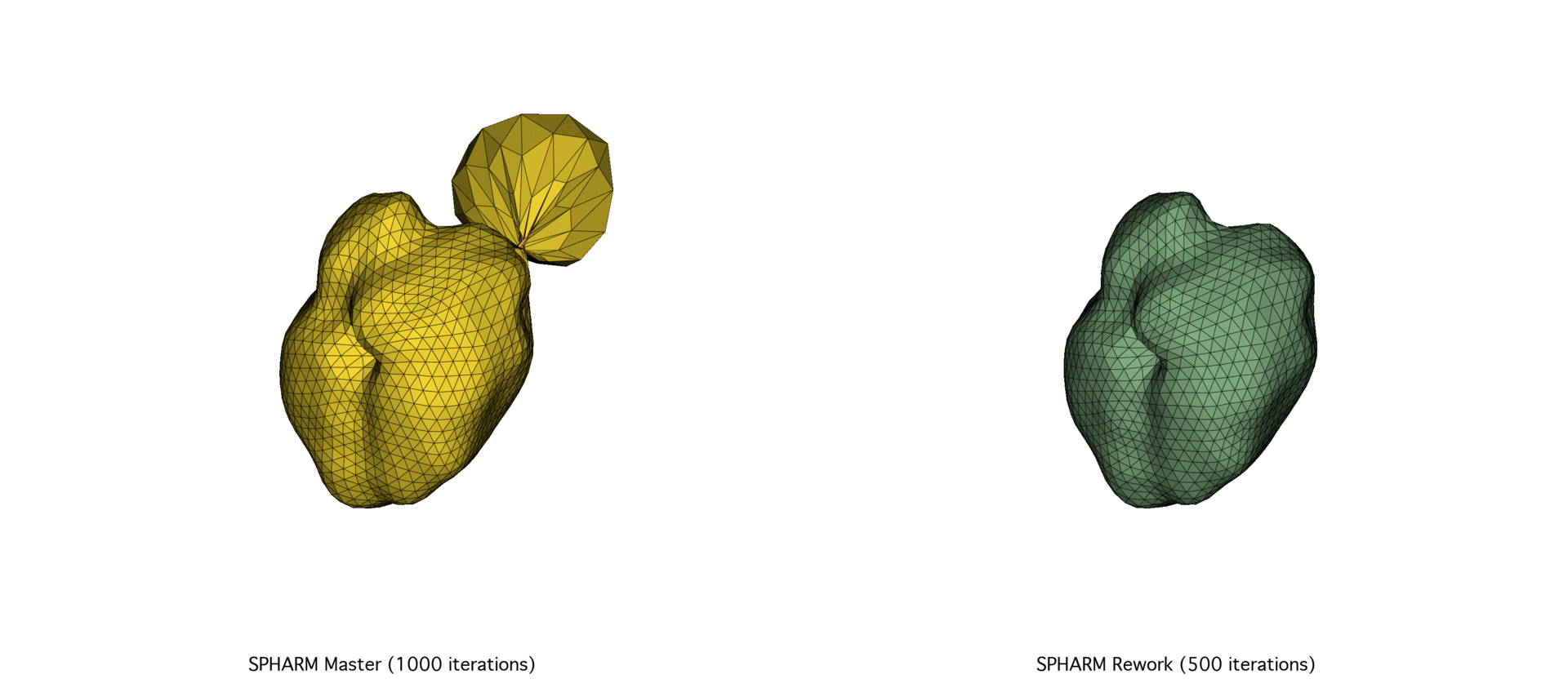
The SPHARM-PDM codebase underwent a major overhaul of its C++ codebase, prompted by the need for better scalability and performance. After exploring a Python port using NumPy and SciPy and facing memory and speed limitations, we focused on optimizing memory allocation and precision tuning in the C++ codebase through Eigen. The reworked version achieved substantial speedups (30–72%) and improved mesh quality, particularly on large datasets.
The updated SurfaceToolbox extension in SlicerSALT 6.0 introduces five new modules. Mesh Alignment, Average Mesh, Surface Distance, Surface Feature Extractor, and Mesh Data Imprinter expand the toolbox’s capabilities from single-model operations to multi-mesh interactions and data exchanges across models, images, and files. The new modules enable tasks such as aligning shapes with or without correspondence, averaging shapes, computing surface distances, extracting and exporting geometric features, and transferring scalar data between models.
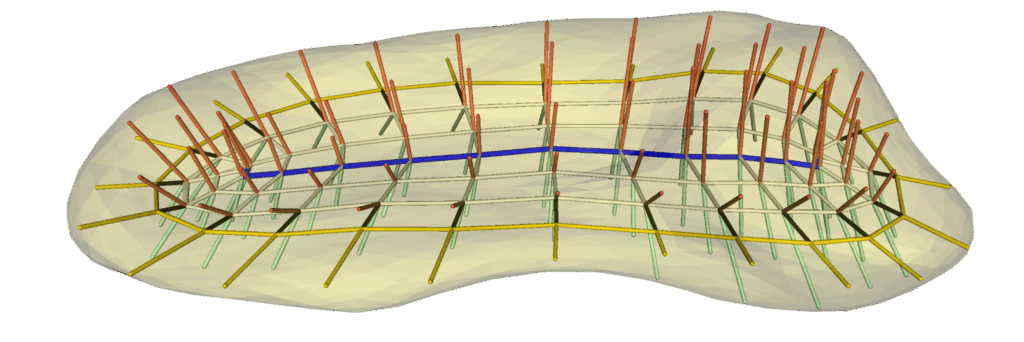
The new evolutionary skeletal representation (s-rep) functionality included in this release uses a similar curvature flow approach to the traditional s-rep fitting already included in SlicerSALT but with several important improvements. For the flow from the input object to an ellipsoid, a conformalized flow is used that has improved geometric properties. For the reverse flow, instead of thin-plate splines we use LDDMM-based registrations via Deformetrica. We also introduce additional geometric constraints so that the crest curve and vertices are mapped more accurately. Results on a hippocampus classification problem show a marked improvement over s-reps fit using previous methods.
The new Difference Statistics module allows for the correlation of shape changes over time between two time points with other clinical variables. It does this by first computing pointwise differences between pairs of corresponding meshes at different timepoints and then using the shape change – instead of the shape itself, along with the given clinical information, through our Covariate Significance Testing module. This results in p-value maps over the object’s surface which show, for each clinical variable, where there is statistically significant correlation with shape changes over time for the given population.
SlicerSALT 6.0 delivers substantial enhancements that strengthen its role as a powerful tool for shape analysis research. These key updates not only improve computational efficiency and usability but also open new possibilities for meaningful insights in biomedical shape analysis. Please visit the SlicerSALT forum for a complete list of improvements and fixes.
Do you want to learn more?
Join our SlicerSALT webinar this coming Wednesday, May 7th at 12pm ET!
During this webinar, we will look into how to compute shape representations, run analysis and visualize results using SlicerSALT. You can register for free here.
Acknowledgements
SlicerSALT is made possible through contributions from a multidisciplinary team coming from Kitware, NYU Tandon School of Engineering and The University of North Carolina at Chapel Hill. The development of SlicerSALT is supported by the NIH National Institute of Biomedical Imaging Bioengineering R01EB021391 (Shape Analysis Toolbox: From medical images to quantitative insights of anatomy).
Get in touch!
SlicerSALT is an example of custom 3DSlicer applications that Kitware develops. Contact us at kitware@kitware.com to learn how we can help you integrate the 3DSlicer-based software into your research, processes and products.