3D Slicer and DICOMweb Networking

In today’s dynamic healthcare landscape, ever-evolving clinical demands call for advanced medical technologies. Medical imaging and health records increasingly rely on data transmission and interoperability. A single CT scan must be readily available for on-site feedback, remote review, cloud-based automated segmentation, and more. The DICOM (Digital Imaging and Communications in Medicine) imaging standard provides a unifying approach to make complex medical images consistently interpretable across a diverse landscape of collection, processing, and viewing software. Similarly, the legacy DIMSE (DICOM Message Service Element) or modern DICOMweb networking protocols provide a common language for communication among medical applications. Picture Archiving and Communication Systems (PACS) act as a central or distributed mechanism for the storage and management of data within a clinical or research environment.
3D Slicer, a powerful open-source platform, offers customizable solutions for medical domains leveraging DICOM and DICOMweb image protocols. This integration enables countless image processing and visualization tools available for use in clinical workflows after regulatory approval. 3D Slicer seamlessly integrates with DICOMweb communications, providing out of the box support while offering room for customizations such as data persistence and authentication settings. DICOM and DICOMweb features in 3D Slicer are made freely available thanks to ongoing collaborative efforts involving key stakeholders and community contributors including Kitware, the Common Toolkit (CTK) groups, the German Cancer Research Center (DKFZ), Isomics, the Perk Lab at Queen’s University, Brigham and Women’s Hospital, the Jolley Laboratory at Children’s Hospital of Philadelphia, and Ascend Cardiovascular. Visit the Slicer ReadTheDocs website for additional contributor information.
In this post, we will walk through a sample workflow to fully understand how 3D Slicer integrates into an image processing workflow with DICOM and DICOMweb support.
Understanding DICOM Data
The first step in processing images is to gain a comprehensive understanding of the available images. The community-contributed “DICOMwebBrowser” Slicer extension provides a turn-key solution to query a PACS server to understand what images are available to load. The extension displays DICOM metadata for visual inspection without actually retrieving each instance.

Alternatively, we can communicate with the remote server directly through the Python interactive console. Slicer bundles the “dicomweb_client” Python module to provide command line DICOMweb support out of the box.
from DICOMLib import DICOMUtils
DICOMUtils.importFromDICOMWeb(
dicomWebEndpoint="https://demo.kheops.online/api",
studyInstanceUID="1.3.6.1.4.1.14519.5.2.1.8421.4009.985792766370191766692237040819")
For scenarios where external and/or offsite servers require authentication for security purposes, Slicer offers a way to configure global DICOMweb authentication parameters from the Slicer command line for reuse across DICOMweb requests.
qt.QSettings().setValue(DICOMUtils.GLOBAL_DICOMWEB_USER_KEY, '<user>')
qt.QSettings().setValue(DICOMUtils.GLOBAL_DICOMWEB_PASSWORD_KEY, '<pwd>')
DICOMUtils.importFromDICOMWeb(
dicomWebEndpoint="https://remote-url/",
studyInstanceUID="1.3.6.1.4.1.14519.5.2.1.8421.4009.985792766370191766692237040819",
auth=DICOMUtils.getGlobalDICOMAuth()
)
In practice, it is common for users to be provided with a set of remote DICOM instances to be retrieved and processed. In the next section, we will look into the process of fetching DICOM data from a remote server.
Fetching DICOM Data
As pictured above, the “DICOMwebBrowser” Slicer extension provides a user-friendly interface for fetching DICOM data from a remote server. Slicer loads DICOM samples into its underlying DICOM database on the local disk, with the option to immediately load samples into the active scene or defer loading until later.
We can customize the DICOM fetch and load behavior programmatically with utilities available in Slicer’s `DICOMLib` Python module. For instance, we can store DICOM images in a temporary local DICOM database that Slicer removes when the data is no longer needed. Additionally, we can configure DICOMweb endpoints and authentication settings either directly from the Python console or within a custom Slicer module.
from DICOMLib import DICOMUtils
with DICOMUtils.TemporaryDICOMDatabase() as db:
DICOMUtils.importDicom(dicomDataDir, db)
Slicer provides a range of mechanisms for loading and interpreting DICOM data such as scalar 2D or 3D volumes out of the box. We can further enhance these capabilities by installing community-contributed Slicer extensions to support extended DICOM standards. For instance, we may load echo ultrasound data with proprietary tags with the SlicerHeart extension.
Manipulating DICOM Data
Once we load DICOM data into a scene we can apply the vast array of image processing tools available within Slicer. These tools apply to a wide range of needs including semi-automated segmentation with MONAI Label and the Slicer Segment Editor module, volume rendering with modality-specific viewing presets, or sequence playback with the Sequences module.
It’s important to note that DICOM images are typically considered immutable, meaning that any updates to the original image data should be stored as new DICOM records. 3D Slicer allows for the conversion for a wide range of results into DICOM records, including basic screenshots, processed volumes, annotations, and even entire scenes.
Results can be converted to DICOM directly from the Slicer Data pane. Most data types can be directly converted to corresponding DICOM storage standards such as for scalar volumes or screenshot secondary captures. We can also save the entire Slicer MRML scene consisting of various in-memory data and scene views as a Medical Reality Bundle (MRB) DICOM secondary capture, which can be easily reloaded into another Slicer scene.
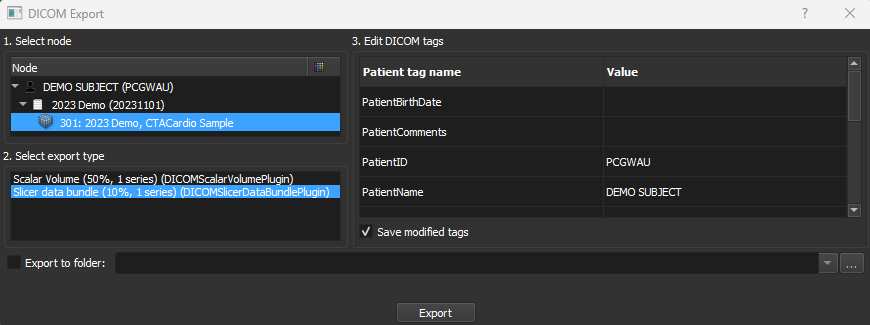
Data exported as DICOM will appear in the Slicer DICOM database.
Storing DICOM Data
We can transmit DICOM records from the active Slicer DICOM database to a remote PACS server directly from the DICOM module user interface. Records can be sent using either the legacy DIMSE or the modern DICOMweb networking protocols.
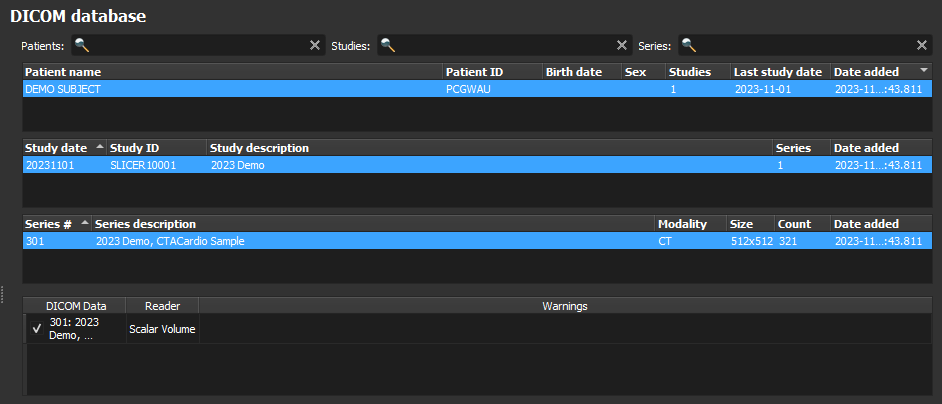
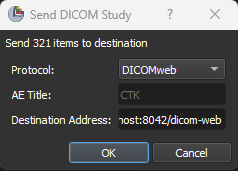
Slicer-based applications can also leverage Slicer’s `DICOMLib` utilities to transmit DICOM records programmatically directly to the PACS system.
from DICOMLib import DICOMSender
sender = DICOMSender(
files=['path/to/0.dcm','path/to/1.dcm'],
address='dicomserver.co.uk:9999'
protocol="DIMSE",
delayed=True
)
sender.send()
In conclusion, the integration of 3D Slicer with DICOM and DICOMweb networking offers a comprehensive solution for processing, manipulating, and transmitting medical images, catering to the dynamic needs of modern healthcare. DICOMweb support enables the integration of 3D Slicer based custom applications into clinical and hospital IT systems for deployment in healthcare settings. This seamless integration empowers medical professionals and researchers to harness the full potential of advanced medical technologies while maintaining data interoperability and security.
Collaborate with Kitware on your 3D Slicer Projects
3D Slicer is fully customizable and a platform of choice for clinical researchers and product developers. Kitware can help tailor the platform to meet your research requirements and incorporate it with your existing tools and workflows. We also provide professional support for 3D Slicer, including training and technical support. Contact our team for more information.