aeva: Annotation and exchange platform for virtual anatomy


We recently announced the release of aeva, a new platform that can represent anatomy in a virtual form for clinical decision making, biomedical research, and medical training. With proprietary, free, and open source software that already exists, researchers commonly use multiple software tools to generate virtual anatomy and prepare it for medical training, additive manufacturing, and computational simulation. For example, image analysis software, such as 3D Slicer, could be used to process medical images, segment organs and tissues, and generate triangulated surface meshes of tissue boundaries. Computer graphics packages like MeshLab and Blender could be used to edit surface meshes, repair, smooth, texture, etc. Meshing software, such as Netgen, TetGen, or Gmsh, could be used for full volumetric discretization of organs and tissues. In addition, working with virtual anatomy is not limited to geometry processing but also requires appropriate and efficient annotation of organs and tissues. Labeling regions to define spatial relationships and interactions between anatomical objects is also critical.
Until now, a unified platform that provides a streamlined interface for the generation, modification, annotation, and exchange of anatomy has not existed. The aeva application suite aims to fill this gap by providing a unified platform with support for diverse data formats and standards. aeva will facilitate in silico description, visualization, and exchange of anatomy between clinicians, scientists, engineers, and the general public. The aeva application suite contains two applications (Figure 1): aevaSlicer and aevaCMB. These two applications jointly provide functionalities that are amenable for the preparation and utilization of anatomy for various workflows (e.g. atlases, machine learning, simulations, etc). These workflows commonly leverage heterogeneous representations of anatomy and require comprehensive descriptions of anatomical relations as frequently seen in physics-based simulations (Figure 2).


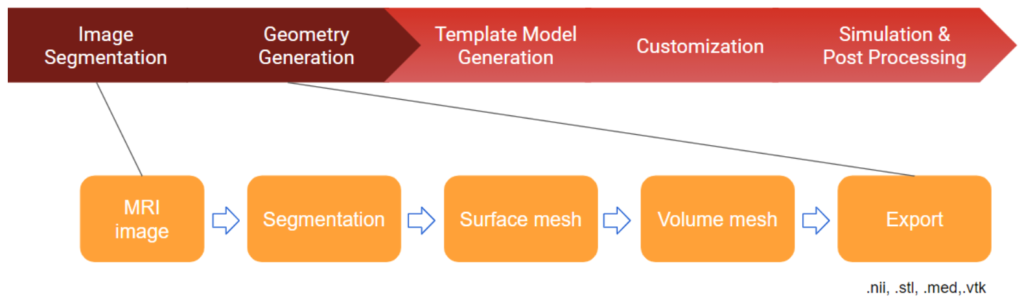
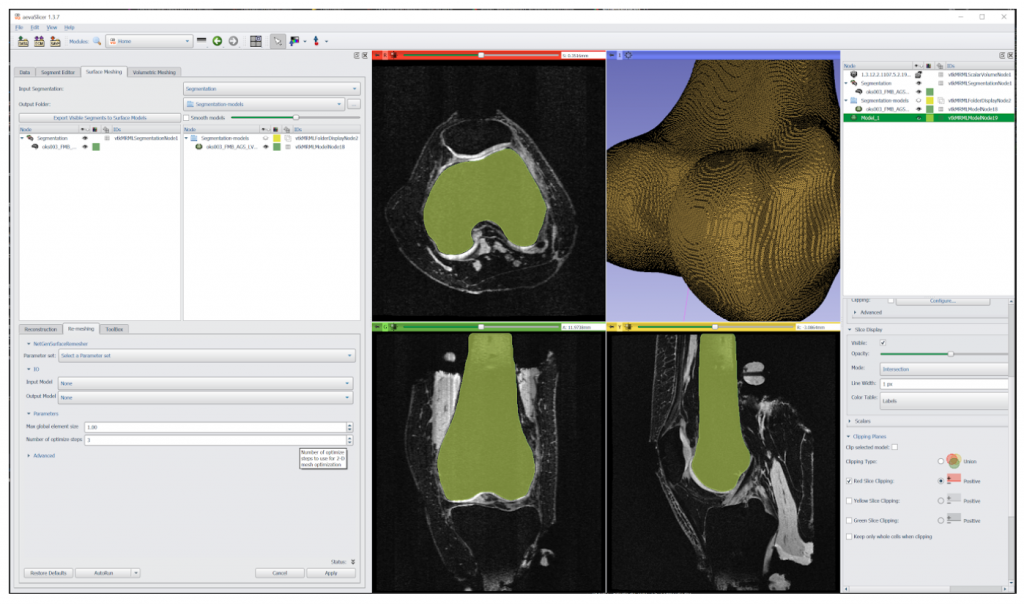
aevaSlicer is designed to provide comprehensive image segmentation, robust surface geometry generation, and a multitude of volumetric meshing capabilities (Figure 3). It is built on top of 3D Slicer—an open source software platform specifically built for medical image informatics, image processing, and three-dimensional visualization. In aevaSlicer, the interface is customized to streamline the workflow to go from an image to a volume mesh of an organ or a tissue (Figure 4). New features have been added to enhance surface generation capabilities and to introduce volumetric meshing of anatomy.
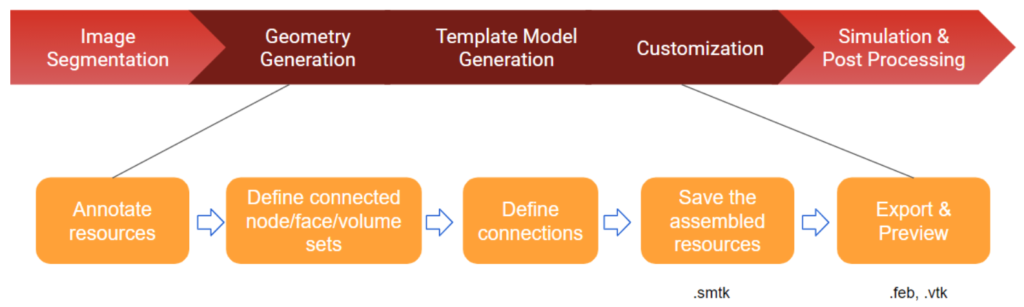
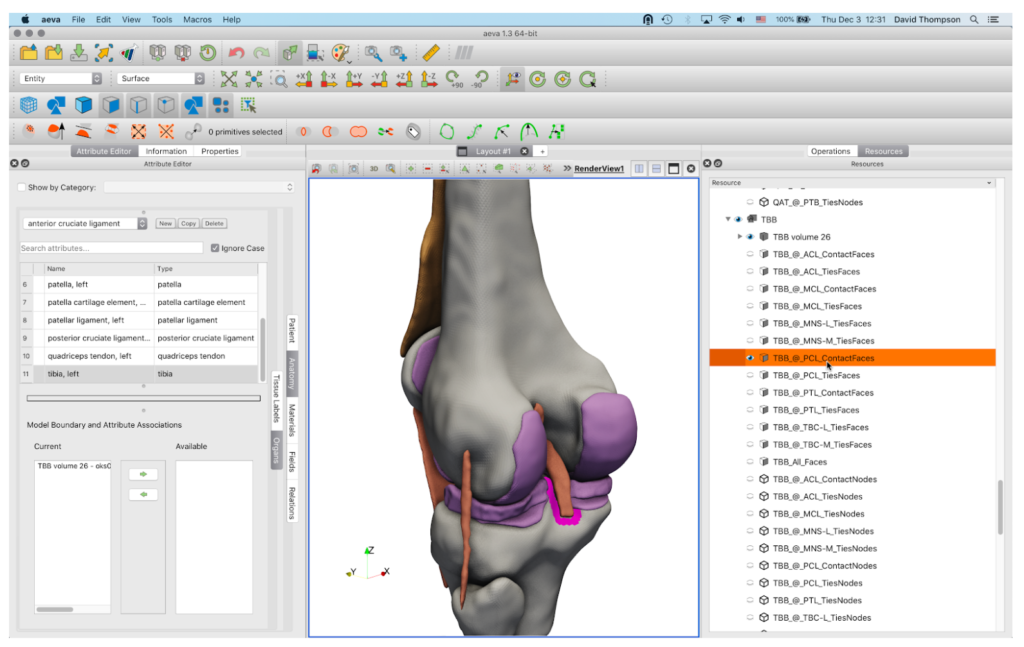
aevaCMB provides resources and templates to assign ontology terms (anatomical or otherwise) to organs and tissues and for appropriate and efficient labeling of regions (i.e. for defining spatial relationships and interactions between anatomical objects), and to generate mesh files that can be loaded to simulation software (Figure 5). aevaCMB is a customized version of CMB—an open source platform for simulation workflows that combines geometric processing with attribute definition, input, and validation to create consistent, high-level descriptions of data appropriate for simulations and other processing. In aevaCMB, the interface is customized for anatomical annotation and representation of mesh data in various formats (Figure 6). New features have been added to support operations for importing and exporting anatomical representations and annotation (template-based and freeform, including a powerful set of region selection tools).
The aeva 2.0 release contains the following key features:
- aevaSlicer
- Streamlined UI
- Surface remeshing
- New surface smoothing operations
- Volumetric meshers (e.g. Netgen)
- aevaCMB
- Capability to load image, mesh, and cad formats
- Templated and freeform annotation (e.g. knee template)
- Robust region selection tools (interactive or automated)
- Native relational storage
- Export to common mesh formats
Please follow the links below to explore the application suite:
- Website: https://simtk.org/projects/aeva-apps (including downloads)
- User guide: https://aeva.readthedocs.io/en/latest/userguide/index.html
- Tutorial: https://aeva.readthedocs.io/en/latest/tutorials/index.html
- Forum: https://simtk.org/plugins/phpBB/indexPhpbb.php?group_id=1767&pluginname=phpBB
- Source Code Repository: https://gitlab.kitware.com/aeva
Acknowledgement
Research reported in this publication was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number R01EB025212. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.