Developing Tools for Exploring Brain Microstructure in Diffusion MRI Datasets
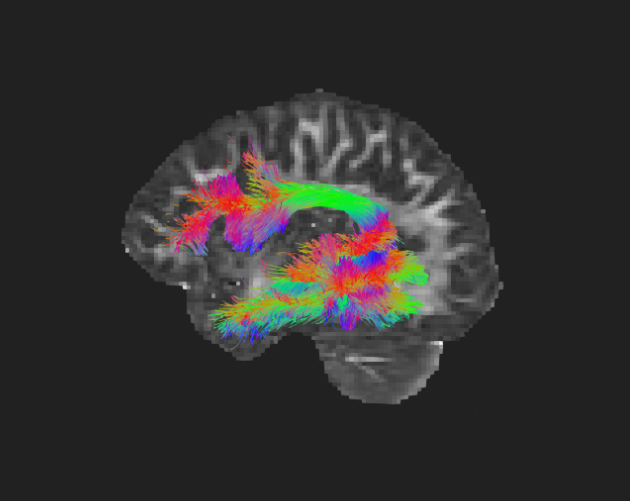
Diffusion MRI is a remarkable neuroimaging modality, simultaneously capturing brain structure at large scales and small. By sensitizing MRI to the diffusion of water molecules, brain tissue structures can be characterized in a directionally-dependent manner at sub-voxel scales.
With microstructure models, diffusion MRI allows us to describe the distribution of neuron arrangements within the imaging voxels. And with tractography, it allows us to infer white matter fiber tracts spanning across regions of the brain. Observing microstructure changes over time provides a window into what goes on during brain development in infancy and adolescence, during life changes, and with the onset of mental illness. As part of an NIH funded project 1R21MH132982, we are building software tools for processing longitudinal population datasets of diffusion MRI and analyzing brain microstructure changes over time. This is a collaboration between Kitware Medical Computing, the UNC Biomedical Image Analysis Group, and the UNC Neuro Image Research and Analysis Laboratories.
This project uses the Adolescent Brain and Cognitive Development (ABCD) Study dataset. The ABCD Study is a large, multi-site, longitudinal study monitoring the brain and cognitive development of a national cohort of children. The dataset includes diffusion MRI data and tabulated data covering a variety of environmental and mental health variables. With the recent 5.1 release, there are currently up to three time points of available imaging per participant, for ages 9-15.
The massive ABCD data covers a wide range of domain areas, including several important aspects of brain development. Many external researchers have used ABCD to tackle their own research questions. However, a secondary analysis does not always hold up to specific research questions as effectively as a smaller and more targeted study. The best way for third party researchers to use the ABCD data for their own investigation is to use it for hypothesis generation [4]. For this reason we have focused our efforts on developing tools for general exploratory analysis rather than hypothesis-driven work. With the exploratory tools that we are putting together, researchers will be able to use the ABCD Study to learn about developmental trajectories in the context of their area of interest, and then they can conduct targeted independent studies to probe the specific hypotheses that they generated.
We aim to provide software that is geared towards processing ABCD diffusion imaging to generate longitudinal trajectories of brain microstructure development. The user of the software will specify how they want to split up groups of study participants and which microstructural features they want to explore. We are taking a Python-plus-Slicer approach to sharing this software: release a standalone Python package with ready-to-use pipelines and a clean API, and release an accompanying 3D Slicer extension that can be used to visualize individual steps and carry out the resulting statistical exploration.
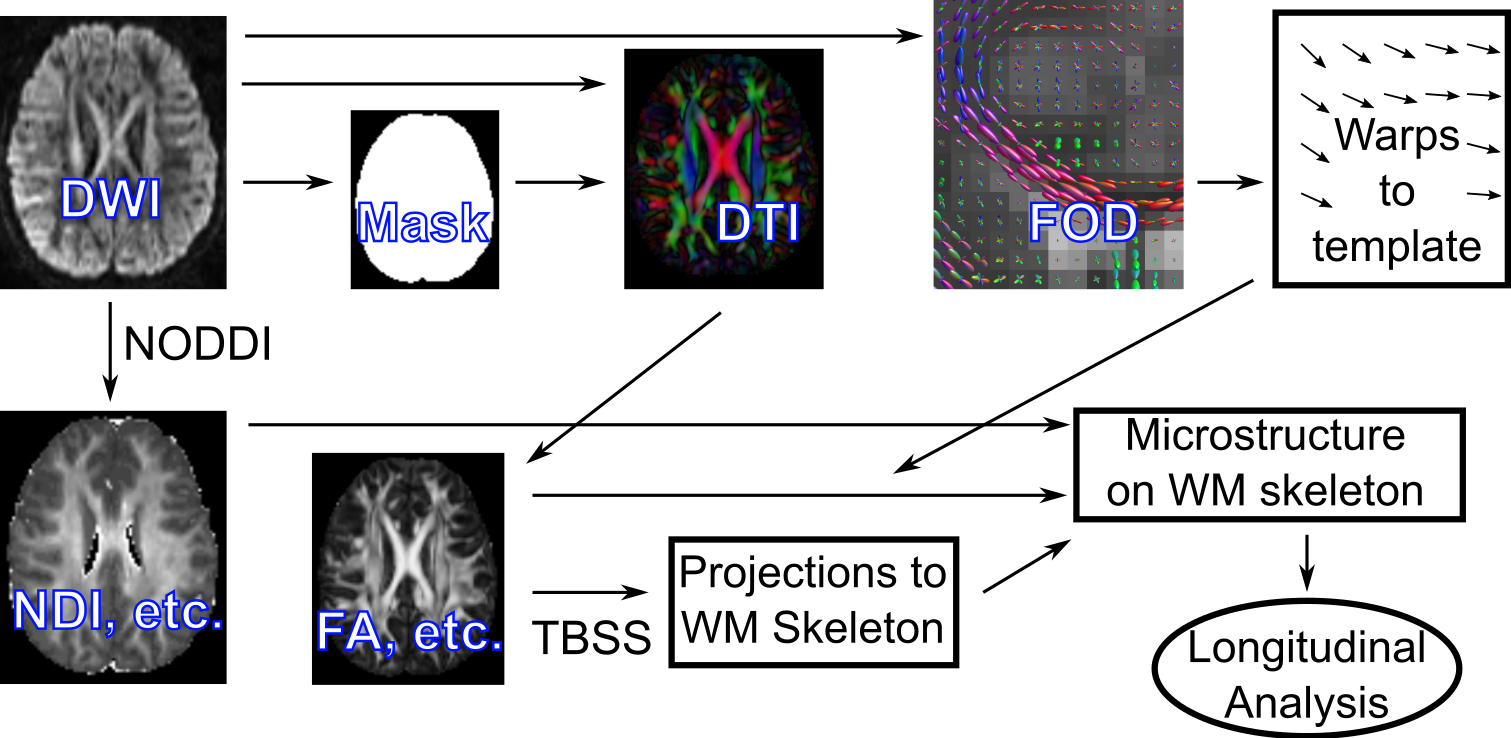
Notable components of the pipeline include data harmonization, image registration, Neurite Orientation Dispersion and Density Image (NODDI) computation, and tract-based spatial statistics (TBSS). We will briefly comment on our approach to each of these items.
Multi-site neuroimaging data contains effects attributable to study site such as scanner hardware, reconstruction algorithms, acquisition protocols, and image quality. These can all affect the final image and downstream results. Data harmonization is the process of removing these effects. In [3], we showed that data harmonization significantly reduces the presence of site-effects in ABCD fractional anisotropy (FA) images, and provide a general-purpose open source tool for image harmonization.
As we will discuss below, fast and accurate registration are critical for our analysis pipeline. There are tools available for fast and accurate registration of scalar images, but we need to ensure accuracy at the level of fiber tract alignment, and this requires a registration algorithm that takes fiber orientations into account. For this reason we have been developing a novel registration methodology that combines deep learning and fiber orientation distributions (FODs).
For deep registration of diffusion images, we elected to train a multi-step, affine and deformable registration algorithm that is inverse-consistent by construction [2]. This approach has several benefits: It is known to work across datasets without hyperparameter tuning, allowing efficient training on our dataset. The combined affine and deformable registration captures large global displacements and detailed local displacements well. Finally, because it is exactly inverse consistent, inverse transforms are directly available and there is never a need to choose which image should be fixed and which should be moving for a particular analysis. To adapt this approach to our diffusion images, we first derive from FOD images with 4 scalar intensities at each voxel corresponding to powers at various spherical harmonic degrees. This representation is rotation-invariant [1], allowing us to use network architectures that were designed to process scalar images. We modify the U-Net and ConvNet components of [2] to have 8 input channels instead of 2 to accommodate two 4-channel images. We compute localized normalized cross-correlation (LNCC) similarity between fixed and warped images across all 4 channels and average. Otherwise, all hyperparameters and training parameters are as in [2].
Since our objective is ultimately to align fiber tracts, we are evaluating and comparing registration methods using metrics that are based on tractography. For each pair of participants in a group (here sampled from ABCD data equally over study sites), we register one subject brain dMRI to the other and then warp fiber tracts along the registration mapping. We then compare fiber tract distances to determine the quality of the registration in terms of fiber alignment. We are still running this evaluation program on a number of dMRI registration algorithms. The outcome will help us decide which algorithm is suitable for the microstructure exploration software, be it our novel fast one or some existing one that performs sufficiently better.
NODDI [7] is the microstructure model we chose to focus on because it is useful for detecting the types of brain changes that occur in adolescent development. One could also use standard measures that are derived from DTI such as FA, and the software can easily be adapted to do this. However FA can be influenced by both neurite density, which changes during adolescence due to neural pruning, and neurite orientation dispersion, which changes during adolescence with reorganization. While FA has the sensitivity to detect these changes in some regions, more advanced diffusion models such as NODDI have the specificity to actually distinguish between them. There are plenty of other choices of advanced diffusion models that are relevant to brain development, but NODDI stands out as both having the right sort of specificity and also having some histological validation [5].
Tract-Based Spatial Statistics (TBSS) [6] is a popular exploratory technique for analyzing DMRI datasets, due to its being automated in readily available software and due to the built-in robustness of the technique to small registration errors. TBSS works by using FA maps from a population dataset to generate a “white matter skeleton” in a common coordinate system. The white matter skeleton is a sort of lower dimensional subspace of the 3D white matter volume (imagine a collection of surfaces and curves, approximated in the form of 3D voxels).TBSS generates a projection map that is typically used to send subject FA values to the skeleton for downstream analysis of clinical groups to be performed on the skeleton voxels. As an automated form of dimensionality reduction that in some sense still respects white matter anatomy, TBSS is excellent for visual data exploration and hypothesis generation. This is why we are working on having TBSS, in addition to the voxel-wise analysis on a population template, as a tool in our exploratory analysis software. In addition to projecting FA values to the skeleton, we will be projecting NODDI indices, and we will be interested in comparing longitudinal trajectories, rather than static values, across groups.
All software for this work, much of which is currently under active development, will be at our github organization brain-microstructure-exploration-tools.
Kitware partners with leading research universities and medical centers, including Cleveland Clinic, the University of North Carolina, and Brigham and Women’s Hospital. Through these partnerships, we create and disseminate high quality software libraries that have a long-term impact on the scientific community.
This work was supported by the National Institutes of Health under Award Number 1R21MH132982. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
[1] Bloy, Luke, and Ragini Verma. “Demons registration of high angular resolution diffusion images.” 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. IEEE, 2010.
[2] Greer, Hastings, et al. “Inverse consistency by construction for multistep deep registration.” International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer Nature Switzerland, 2023.
[3] Osika, Tom, et al. “Harmonization Benchmarking Tool for Neuroimaging Datasets.” 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI). IEEE, 2023.
[4] Saragosa-Harris, Natalie M., et al. “A practical guide for researchers and reviewers using the ABCD Study and other large longitudinal datasets.” Developmental cognitive neuroscience 55 (2022): 101115.
[5] Schilling, Kurt G., et al. “Histological validation of diffusion MRI fiber orientation distributions and dispersion.” Neuroimage 165 (2018): 200-221.
[6] Smith, Stephen M., et al. “Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data.” Neuroimage 31.4 (2006): 1487-1505.
[7] Zhang, Hui, et al. “Axon diameter mapping in the presence of orientation dispersion with diffusion MRI.” Neuroimage 56.3 (2011): 1301-1315.