Integrating AI and Data Science in Digital Pathology
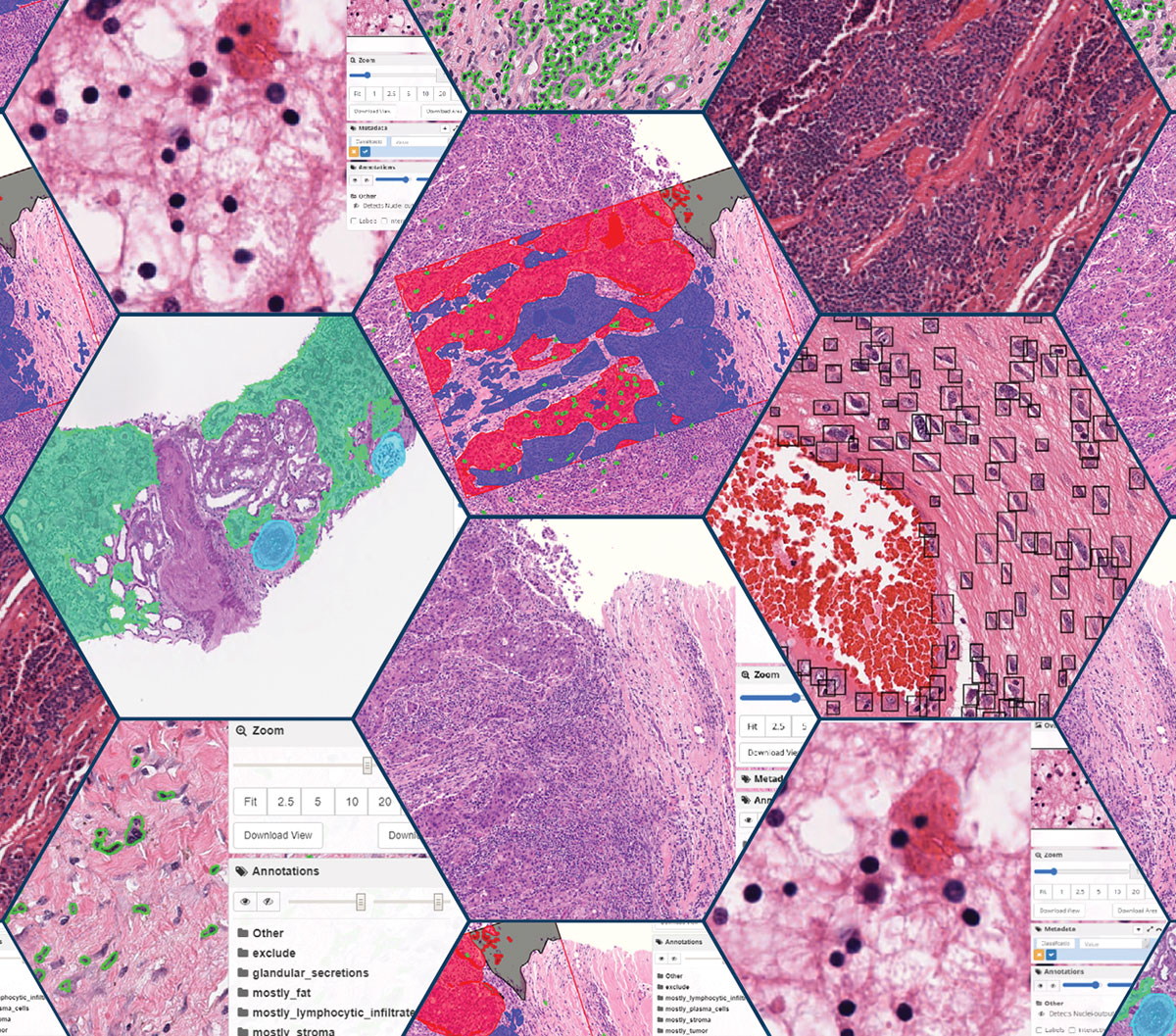
Artificial intelligence (AI) and machine learning (ML) are revolutionizing digital pathology, enabling more precise analysis and accelerating research and clinical workflows. However, integrating these technologies effectively requires overcoming challenges in data management, model deployment, and workflow customization.
Key Challenges in Digital Pathology AI
Effective AI integration requires addressing multiple technical hurdles:
- Data management: Pathology datasets are large, multi-format, and often distributed across institutions. Organizing, filtering, and accessing these datasets efficiently is critical.
- Annotation quality: Comprehensive, high-quality annotations are essential for robust model training and reliable results.
- Model deployment: Solutions must run seamlessly on local machines, cloud platforms (AWS/GCP), or high-performance computing (HPC) environments.
- Workflow customization: Different pathology tasks demand tailored user interfaces, pipelines, and processing strategies.
HistomicsTK, Kitware’s open source digital pathology platform, is designed to overcome these obstacles, enabling teams to focus on insights rather than infrastructure.
HistomicsTK: A Platform for Scalable AI Integration
HistomicsTK provides a modular, open source framework that supports the full AI workflow in digital pathology:
- Flexible data access and organization: Store data centrally or access it in place from S3 buckets or other institutions. Organize datasets into studies for training, testing, and validation, with customizable views based on metadata.
- Comprehensive annotation tools: Annotate WSIs with heatmaps, grids, images, and shapes; support multi-channel fluorescence images; and integrate annotations from GeoJSON, Aperio XML, and other formats.
- Algorithm integration and execution: Run AI/ML algorithms via the Slicer Execution Model, with containerized support (Docker, Podman, Apptainer) and batch processing for scalability.
- Customizable interfaces and workflows: Optimize UIs for specific tasks, create guided labeling pipelines, and extend functionality with custom plugins.
- Flexible deployment: Operate locally, in the cloud using Terraform, or on HPC systems with independent worker nodes.
Check out our webinar recording with a live demo to learn more about these workflows in action.
Real-world applications of HistomicsTK
- Guided Labeling with Northwestern University: Pathologists efficiently labeled superpixels to train custom AI models, demonstrating scalability across diverse annotation types.
- BANFF-AID: Automating Kidney Biopsy Analysis: AI-powered segmentation and lesion scoring for glomeruli, tubules, and arteries reduced variability in manual assessments, illustrating how models can improve diagnostic consistency.
- Foundational Model Integration: Simple workflows demonstrated how new AI models can be rapidly applied to existing datasets for evaluation and comparison.
Additionally, open source extensions such as MONAI Label and ImageDePHI allow teams to leverage pre-trained models and anonymize sensitive datasets, respectively.
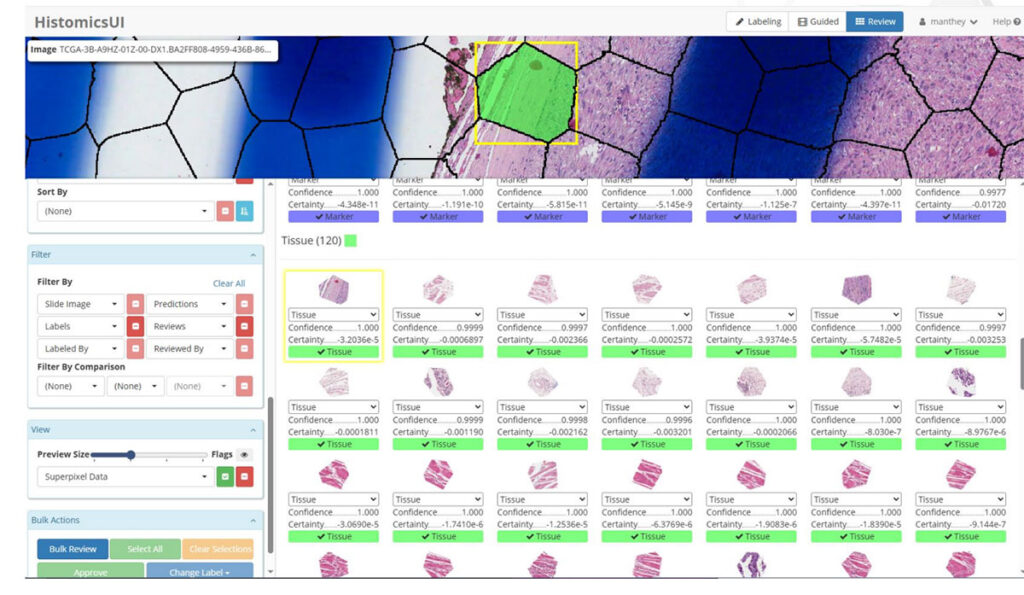
Partnering with Kitware for AI-Driven Pathology
With decades of experience in open source software and scalable scientific computing, Kitware built HistomicsTK to make AI integration practical and approachable. Whether you’re handling massive pathology datasets, training custom AI models, or running workflows on local machines, the cloud, or HPC systems, HistomicsTK gives you the flexibility and tools you need, so you can focus on discoveries, not infrastructure.
If you’re interested in exploring how HistomicsTK can accelerate your AI-driven pathology projects, our team can provide technical guidance, collaboration opportunities, and real-world examples.
Contact us to get started.