Integrating NVIDIA Clara Models into VolView: A Technical Deep Dive
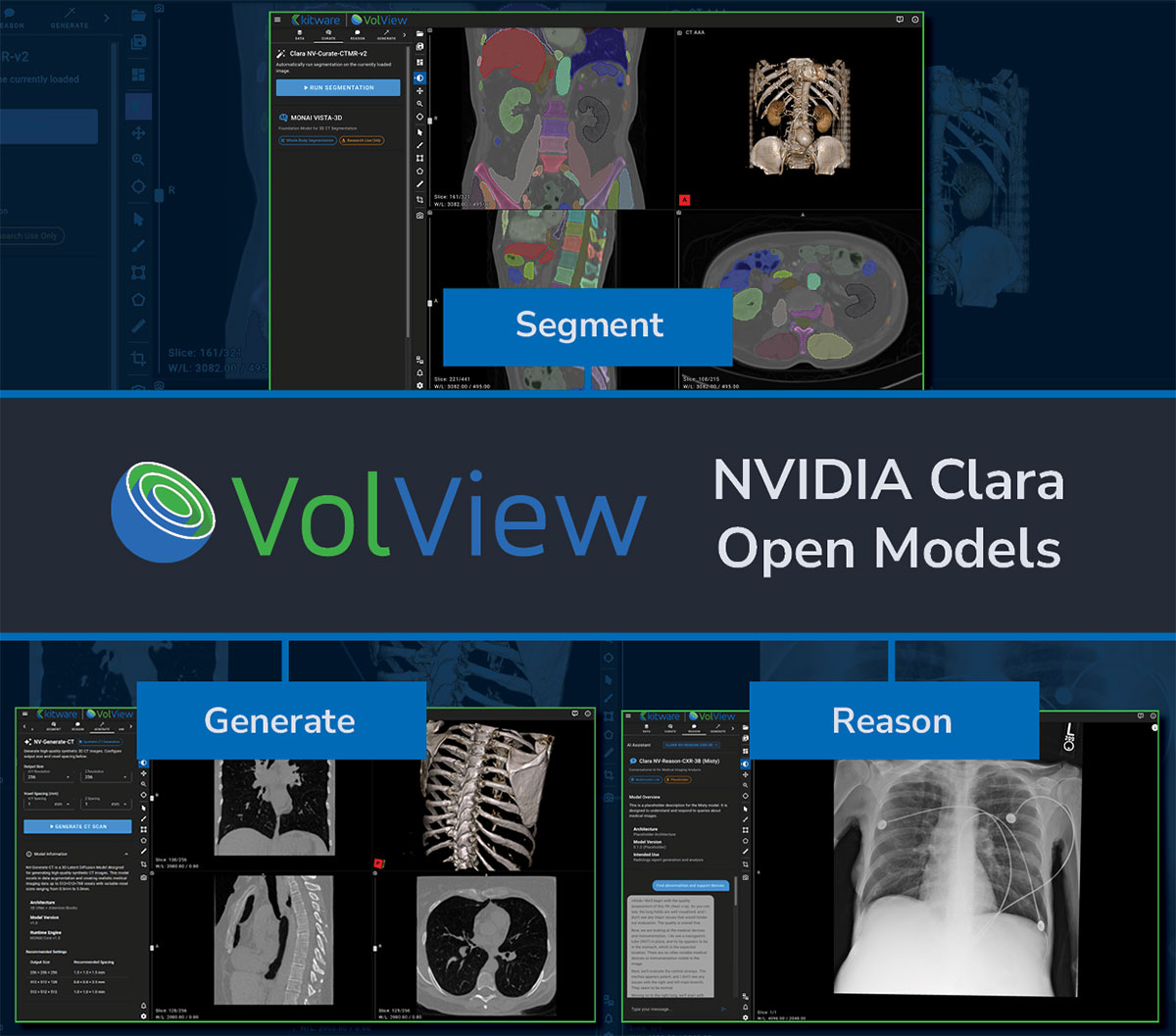
Introduction
This post is a technical deep dive into the integration of Clara open‑source models into VolView’s browser-native imaging platform. We explain the architecture, design choices, and deployment patterns that bring segmentation, reasoning, and synthetic data generation directly into the browser with zero client installation.
VolView
VolView is Kitware’s zero-install, browser-native medical imaging platform that brings workstation-class visualization directly into a web browser. Unlike traditional imaging applications that require complex installation procedures, specialized hardware, or remote rendering servers, VolView runs entirely on the client. This means all interaction scrolling through slices, adjusting window/level, cinematic volume rendering, multiplanar reconstruction happens locally on the user’s device with immediate responsiveness.
At the core of VolView is a modern web technology stack built for high-performance scientific visualization. WebGL and WebGPU provide GPU-accelerated rendering directly in the browser, while VTK.js powers interactive 2D and 3D visualization capabilities rooted in decades of Kitware’s experience developing VTK, ParaView, and other large-scale visualization systems. For image processing, ITK-WASM delivers fast, in-browser operations for reading, decoding, resampling, measuring, and manipulating medical volumes using WebAssembly. Together, these components allow VolView to support a wide array of formats including DICOM, NIfTI, NRRD, and MHA without plugins or backend servers.
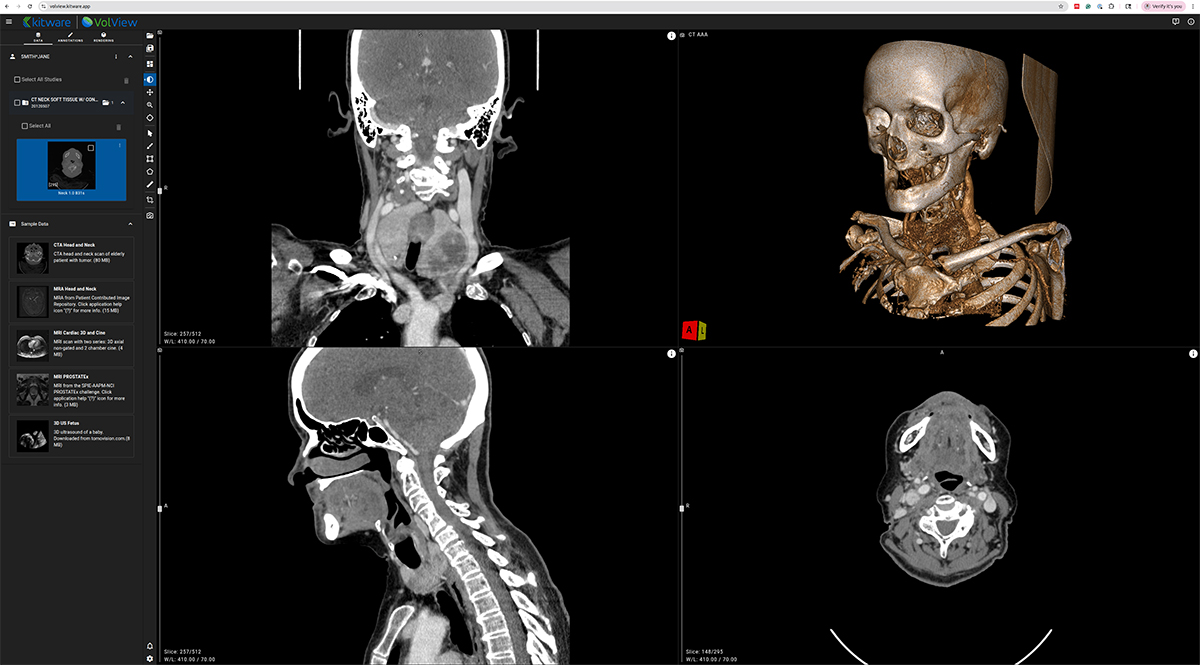
VolView is also built to be simple to adopt and easy to extend. As a fully open-source project, organizations can tailor its interface, visualization tools, or file workflows to their specific needs. Clinicians can use it as a lightweight viewer; researchers can use it for volumetric exploration; developers can embed it into larger web platforms or clinical prototypes. Its modular design and standards-based foundations make VolView suitable for education, research, telemedicine scenarios, and any setting where accessibility and high-quality visualization are essential. This architecture gives clinicians, researchers, and developers access to advanced visualization without installing software, provisioning GPU workstations, or maintaining a streaming infrastructure. As the ecosystem shifts toward browser-native scientific computing, VolView demonstrates what is now possible with modern web technology.
AI Integration Architecture Overview — In Depth
VolView’s architecture is intentionally decoupled to combine a fast, client‑side user experience with scalable AI services. At a high level, there are four layers: (1) data sources, (2) the browser‑based viewer, (3) a lightweight communication layer, and (4) backend services for AI inference and workload orchestration. This layered structure ensures that each part of the system can evolve independently: the viewer can improve without changing backend services, and models can be upgraded without modifying the client application.
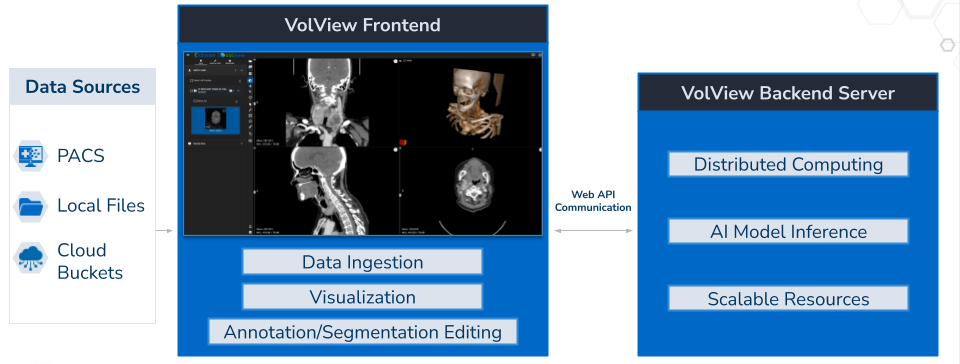
- Data Sources: VolView reads from PACS, local files (drag‑and‑drop), and cloud buckets. Because all visualization is client‑side, users can interactively explore studies with low latency even when data is remote.
- Browser‑Based Viewer: The frontend is built with modern web technologies (TypeScript/Vue, VTK.js for 2D/3D visualization, and ITK‑WASM for in‑browser image processing and DICOM handling). Cinematic volume rendering and common radiology tools (window/level, measurements, annotations) are available out of the box. Crucially, the rendering pipeline runs in the user’s browser with no remote rendering or thick client install.
- Communication Layer: When an action requires server resources—e.g., launching a segmentation or reasoning job—the viewer sends a small API request and subscribes to results via WebSockets. This keeps the UI responsive; model outputs stream back as they are produced and are immediately overlaid on the image canvas for review and editing.
- Backend Services (Segment, Generate, Reason): Each AI capability is provided by an independent service. Services can be scaled horizontally and deployed on any NVIDIA‑enabled GPU infrastructure: a single workstation, a GPU cluster, or a managed cloud.
The detailed system architecture diagram below maps each layer of VolView’s technology stack from the browser rendering engine to the backend microservices, message broker, and storage systems showing the concrete components used in real deployments
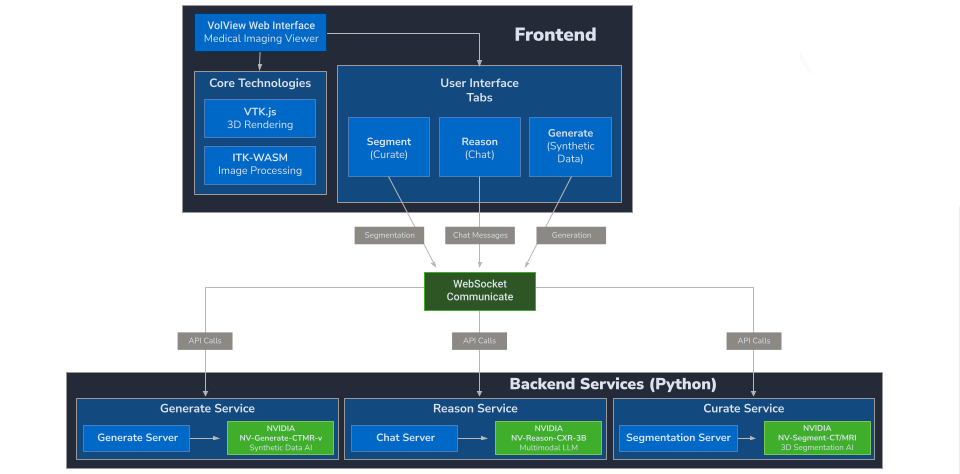
New models can be added by implementing the service interface (REST/WebSocket endpoints plus a simple job contract). Because models are isolated behind service boundaries, you can iterate on architectures, weights, and acceleration (e.g., PyTorch/CUDA, TensorRT, ONNX Runtime) without touching the viewer.
Clara Model Integration: Segment, Generate, Reason
VolView integrates three of NVIDIA’s newly released open-source Clara models—each designed to support a different aspect of medical imaging workflows. Rather than covering the full technical depth of these models here, we focus on how they fit into VolView’s user experience and provide links for readers who want to explore the underlying research.
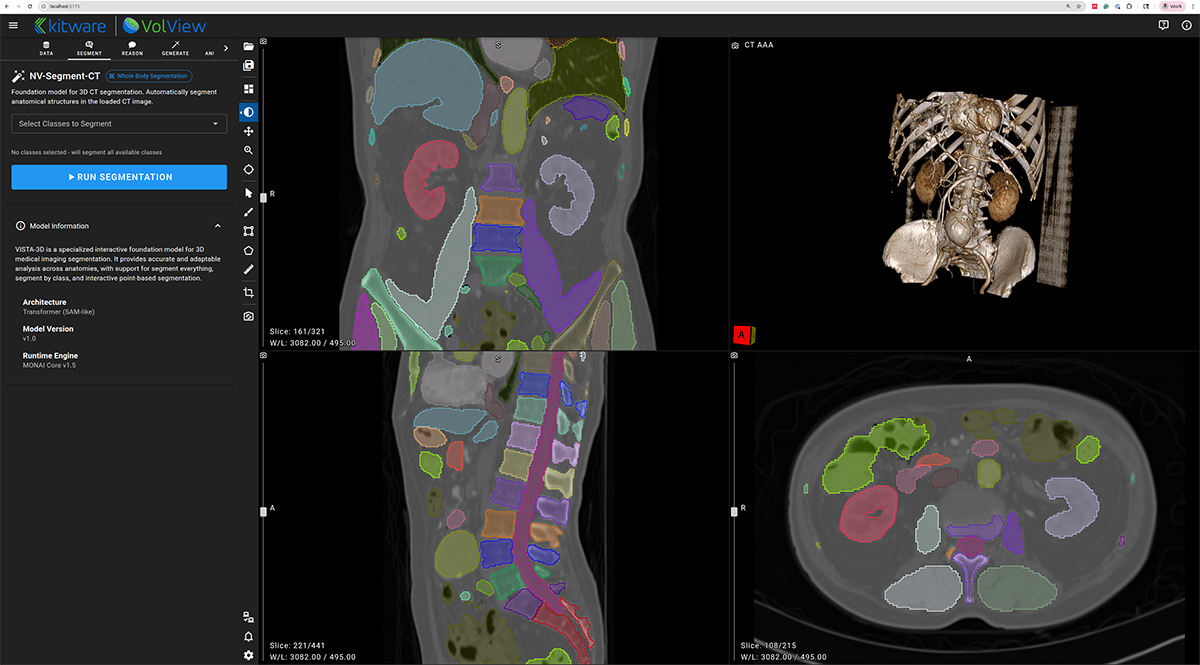
NV-Segment-CTMR — 3D Foundation Segmentation
NV-Segment provides high-quality automatic and interactive segmentation for a wide range of anatomical structures. Within VolView, it enables users to quickly generate volumetric labels, refine results through interactive adjustments, and incorporate segmentation into downstream visualization or analysis workflows.
For technical details, see the paper:
[1] https://arxiv.org/pdf/2406.05285
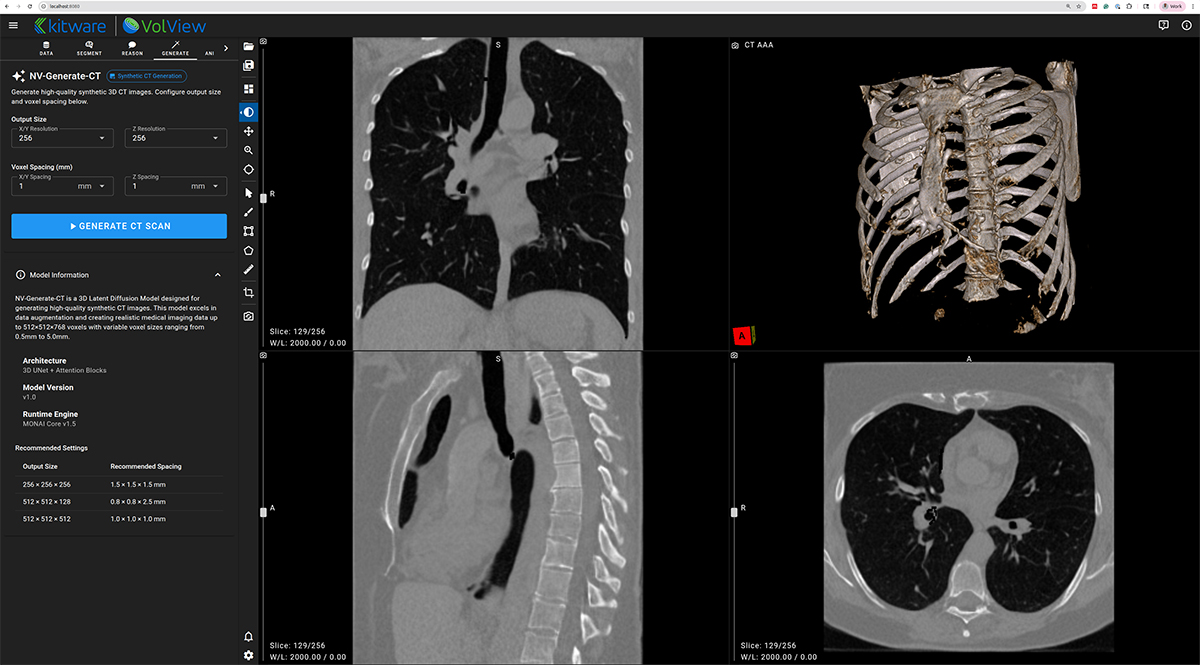
NV-Generate-CT/MR — 3D Synthetic CT and MR Generation
NV-Generate introduces the capability to generate realistic, anatomically consistent synthetic CT and MR volumes. In VolView, NV-Generate supports use cases such as data augmentation, educational content creation, and prototyping scenarios where non-protected health information (PHI) synthetic data is essential.
Learn more in the published work:
[2] https://arxiv.org/html/2409.11169v1
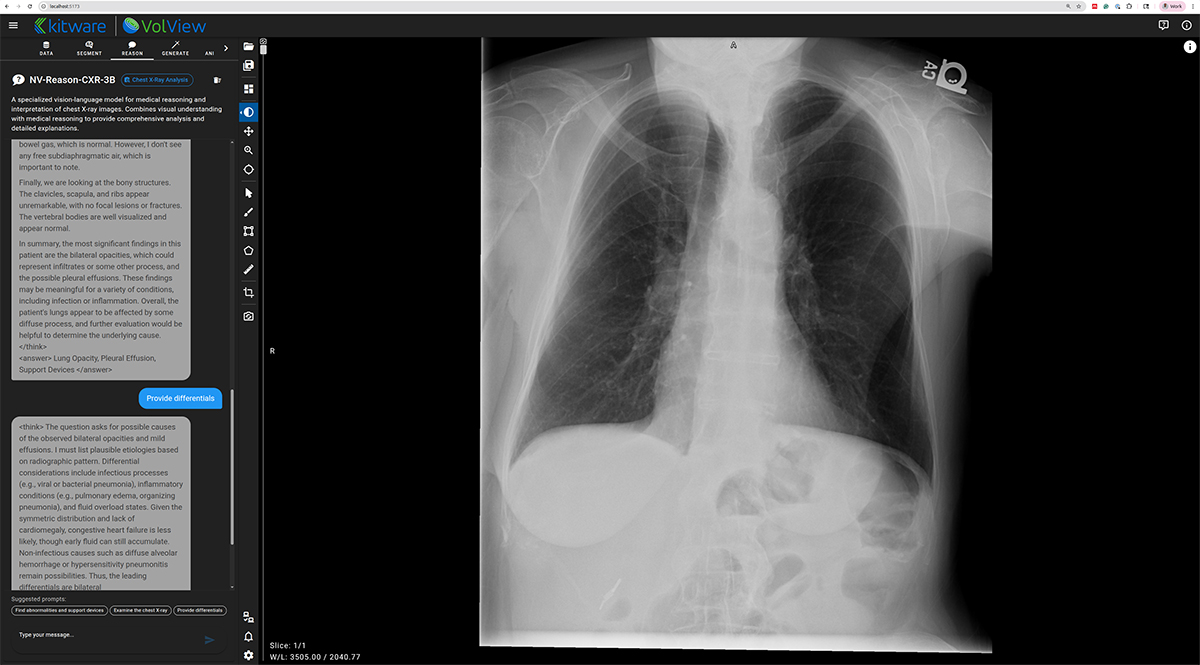
NV-Reason-CXR-3B — Clinically Aligned Chain-of-Thought Reasoning
NV-Reason powers an interactive, multimodal chat interface inside VolView, allowing users to ask questions directly about an image and receive structured, interpretable reasoning. This creates opportunities for explainability, teaching, quality assurance, and exploratory research.
More information is available here:
[3] https://arxiv.org/abs/2510.23968
Watch the Webinar — Full Walkthrough & Live Demo
For a deeper guided walkthrough, the recorded webinar provides a complete end-to-end demonstration. It covers the motivation behind client‑side rendering, an architecture overview, and a live demo of Segment, Generate, and Reason in VolView, along with model validation considerations and pathways to adapt these workflows for institutional requirements.
During the demo, we show whole-body organ segmentation overlays, interactive correction, multimodal reasoning on chest X-rays, and the generation of high-fidelity synthetic CT volumes. The session concludes with deployment guidance and a Q&A session on customization, cloud scale-out, and governance.
The webinar pairs the architectural explanation with a practical demonstration, making it the best starting point for teams evaluating VolView for clinical research, prototyping, education, or workflow integration.
Next Steps
Whether you’re prototyping new research ideas, deploying on-prem clinical tools, or exploring what’s possible with open models, VolView’s architecture gives you a flexible foundation to move quickly and scale with confidence. To dive deeper, we encourage you to launch the demo and explore the codebase to see how imaging, client-side rendering, and AI services come together in a unified workflow:
https://github.com/KitwareMedical/volview-gtc2025-demo
From there, you can integrate your own models through the lightweight service interface or pilot a small deployment on a single GPU before expanding into a larger on-prem or cloud environment. And if you’re developing custom workflows, adding new capabilities, or planning a production deployment, Kitware’s team is available to collaborate on architecture, integration, and scaling strategies tailored to your needs.
We’d love to hear your feedback and partner with teams advancing imaging, AI, and clinical innovation—feel free to reach out anytime.