ITK 5.3 Release Candidate 4 available for testing
We are happy to announce the Insight Toolkit (ITK) 5.3 Release Candidate 4 is available for testing! :tada: ITK is an open-source, cross-platform toolkit for N-dimensional scientific image processing, segmentation, and registration.
ITK 5.3 is a feature release that accelerates performance, provides new segmentation and shape analysis algorithms, improves documentation, adds distributed computing support, among many more improvements. For more information about performance improvements, see the 5.3 RC 1 Release Notes. For more information about new segmentation and shape analysis algorithms, see the 5.3 RC 2 Release Notes. For more information about documentation improvements, see the 5.3 RC 3 Release Notes.
ITK 5.3 RC 4 highlights distributed computing support with Dask. Dask is a Python library that makes scaling analysis easy through simple programming on a laptop that can then be deployed to HPC or cloud computing resources. In ITK 5.3 RC 4, Dask support applied in medical imaging, bioimaging, and material science, is robust (caveat: import itk should be called in Dask worker functions). Furthermore, support was expanded from NumPy array views on itk.Image‘s to full metadata-perserving distributed computing with itk.Image, itk.Mesh, itk.PointSet, and itk.Transform. With ITK’s Dask support, batch processing a cohort of thousands of medical images or processing biomicroscopy, histopathology, or geospatial images with trillions of pixels is now a matter of minutes instead of weeks.
ITK 5.3 RC 4 also includes advancements in Python interface file (.pyi) support and new remote modules to build WebAssembly processing pipelines to native executables and support ITK WebAssembly file formats, perform multimaterial tetrahedral meshing from segmentations, and read meshes from SWC files, a format for representing neuron morphology.

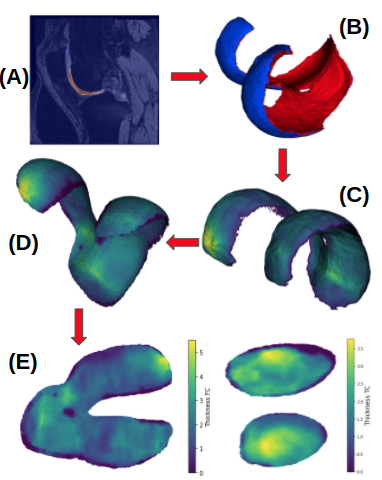
Knee MRI mapping of cartilage thickness in osteoarthritis that leverages ITK’s Dask support for distributed processing of large patient cohorts over the preprocessing, segmentation, registration, and post-processing steps of the analysis pipeline.
Download
Python Packages
Install ITK Python packages with:
pip install --upgrade --pre itkGuide and Textbook
Library Sources
Testing Data
Unpack optional testing data in the same directory where the Library Source is unpacked.
Checksums
Features
Python
- Python packages now include oneTBB support for improved performance
- Following CPython’s deprecation schedule Python 3.6 is no longer supported
- Python packages added for Python 3.10
- Initial Python wrapping is available for the Video modules
TransformToDisplacementFieldis now available in Python- Pythonic IO functions
itk.imreadunderstandspathlib.Path‘s - New
reprforitk.Matrix np.asarrayworks onitk.MatrixDCMTKImageIOwrapping addressedGradientDifferenceImageToImageMetricwrappedSynImageRegistrationMethod,BSplineSynImageRegistrationMethodwrappedConjugateGradientLineSearchOptimizerv4wrapped- Wrap
ImageRegistrationMethodv4foritk.Mesh - Wrap
PointSetToPointSetMetric,PointSetToPointSetRegistrationMethod - Wrap
ANTSNeighborhoodCorrelationImageToImageMetricv4 - Nearly all registration v4 classes are now wrapped
VectorImageinput forDisplacementFieldTransform- Python wrapping for spatial orientation functionality
- PyImageFilter wrapped for additional types, supports pipeline functionality
- NumPy array interfaces for
itk.PointSet,itk.Mesh - manylinux_2_28 and manylinux2014 wheels are provided
- Dask support for
itk.Image,itk.PointSet,itk.Mesh,itk.Transform
C++
- C++14 is now required
- The minimum CMake version required is now 3.16.3
- New functions:
MakePoint,MakeVector,MakeIndex,MakeSize. - Targets in Visual Studio and other IDE’s are now organize hierachically by ITK Group and Module
- Most of
itk::mplmeta-programming functions replaced by C++14 equivalents - Performance accelerations for b-spline interpolation, Mattes mutual information metric computation
- Improved modern C++ adoption, e.g. additional adoption of
constexpr,auto itk::ReadMesh,itk::WriteMeshsimple reader functions available, similar toitk::ReadImage,itk::WriteImage- FFT backends are now registered through the object factory mechanism
cbegin()andcend()member functions toIndex,Offset,Size- Add
itk::MakeFilled<TContainer>(value) itk::ConvertNumberToString<TValue>(val)convenience functionitk::bit_cast<TDestination>(source)functionitk::PolyLineCellInputSpaceNameandOutputSpaceNamesupport foritk::Transformqfac,qt_xyzadded to Nifti metadataLZWcompression support- Support requested output region in FFT filters
- Many code coverage improvements
New filters
itk::TransformGeometryImageFilter: applies a rigid transform to anImage‘s metadata.- 1D FFT classes
- Interface classes for forward, inverse transformations
- Vnl implementations
- FFTW implementations
itk::TriangleMeshCurvatureCalculator– Gaussian curvature calculator foritk::MeshFFTDiscreteGaussianImageFilter— discrete gaussian filters via FFTs
Remote module updates
New remote modules:
- HASI: High-Throughput Applications for Skeletal Imaging
- ITKGrowCut: segments a 3D image from user-provided foreground and background seeds
- ITKMeshToPolyData: Convert an ITK Mesh to a simple data structure compatible with vtkPolyData
- ITKCudaCommon: Framework for processing images with CUDA
- itk-wasm WebAssemblyInterface: Build WebAssembly processing pipelines to native executables and support ITK WebAssembly file formats
- ITKCleaver: Multimaterial tetrahedral meshing.
- ITKIOMeshSWC: Read meshes from SWC files, a format for representing neuron morphology.
Updated modules: AdaptiveDenoising, AnisotropicDiffusionLBR, BSplineGradient, BoneEnhancement, BoneMorphometry, Cuberille, GrowCut, HASI, HigherOrderAccurateGradient, IOFDF, IOScanco, IsotropicWavelets, MinimalPathExtraction, Montage, MorphologicalContourInterpolation, RTK, SimpleITKFilters, SkullStrip, SplitComponents, Strain, TextureFeatures, Thickness3D, TotalVariation, TubeTK, and Ultrasound.
Third party library updates
- dcmtk
- eigen
- expat
- fftw
- gdcm
- googletest
- hdf5
- kwsys
- kwiml
- minc
- metaio
- niftilib
- vxl
- zlib migrated to zlib-ng
Detailed Changelog
Find a detailed changelog in the full release notes.
Congratulations
Congratulations and thank you to everyone who contributed to this release.
Of the 76 authors who contributed since v5.2.0, we would like to specially recognize the new contributors:
Michael Kuczynski, Tim Evain, Tomoyuki SADAKANE, Mario Emmenlauer, Andreas Gravgaard Andersen, Ebrahim Ebrahim, josempozo, Wenqi Li, Genevieve Buckley, Oleksandr Zavalistyi, Jose Tascon, Pranjal Sahu, ambrozicc1, Vagrant Ca scadian, MrTzschr, Philip Cook, Tihomir Heidelberg, Jason Rudy, Kian Weimer, z0gSh1u, Darren Thompson, Darren, Jose M Pozo, Paul Elliott, Gabriele Belotti, Rafael Palomar, Fernando Hueso-González, Mark Asselin, mrhardisty, Laryssa Abdala, Roland Bruggmann, Natalie Johnston, and ferdymercury.
What’s Next
This is the last release candidate before the 5.3.0 release. Please try out the current release candidate, and discuss your experiences at discourse.itk.org. Contribute with pull requests, code reviews, and issue discussions in our GitHub Organization.
Enjoy ITK!