MONAI and ITKWidgets: Getting Started
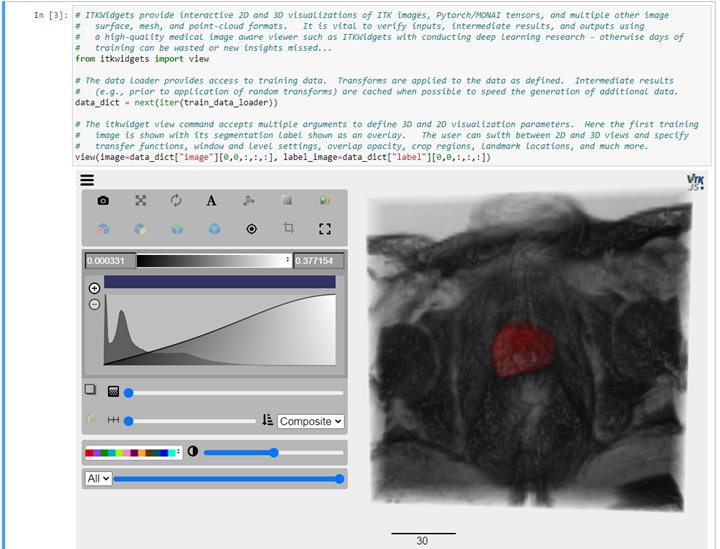
This is our curated list of high-level concepts, code examples, course material, and web resources for getting started with MONAI and ITKWidgets.
MONAI is a freely available, open source, deep learning library, based on PyTorch and specialized for medical image analysis. ITKWidgets brings interactive 2D and 3D visualization and annotation capabilities to Jupyter Labs and Notebooks.
Together, MONAI and ITKWidgets enable deep learning inputs, intermediate values, and results to be visualized, explored, and optionally refined by researchers and clinical collaborators with ease. Such interactive visualizations of inputs and intermediate results are crucial to avoiding days of wasted training and to gaining new insights into results and future research directions.
The material presented in this blog is meant as a getting-started guide for a wide range of users and developers. Some knowledge of Python is assumed; however, even those with no python experience should benefit from much of the material presented herein.
Why MONAI?
MONAI began as a collaboration between KCL and Nvidia but rapidly expanded to include a massive community of highly skilled and very dedicated researchers and developers from around the world. Why was this adoption so quick? Because MONAI addresses the specific needs and opportunities of deep learning in medical imaging:
- Medical Image Data: MONAI supports a variety of medical image formats (thanks to ITK, GDCM, NIBabble, and other toolkits), and it respects the physical properties of those images, e.g., pixel size, pixel spacing, and image orientation. Those physical properties are often ignored by traditional computer vision libraries, but they are essential to the accurate and real-world application of deep learning to clinical medical images.
- Medical Image Transforms: MONAI includes a multitude of domain-specific transforms for medical image pre-processing and augmentation. Unlike with photographs or other common computer vision data, it is often not appropriate to rotate, flip, or change the intensities (e.g., Hounsfield Units) of medical images for augmentation purposes, yet warping the anatomy in images to match patient variations, removing ribs from x-ray images, and other steps are common in medical image pipelines and are provide by MONAI via ITK, DKFZ’s BatchGenerator, ICL’s DLTK, KCL’s NiftyNet, and other transform libraries that have been integrated into MONAI.
- Medical Workflows: 3D Slicer provides a user-friendly interface for applying trained MONAI models to clinical workflows for a variety of disease diagnosis, treatment planning, and treatment guidance research. Similarly, AWS’s SageMaker, Google’s CoLab, and Nvidia’s Clara provide cloud-based platforms for hosting MONAI research, development, and deployment platforms. Efforts to integrate MONAI inference capabilities with easy-to-deploy web-based 3D radiological viewers such as ParaViewMedical are also underway.
- Reproducibility and Standardization: Underlying the design and implementation of MONAI is a commitment to promote reproducibility and build upon existing standards. Reproducibility is paramount to clinical deep learning regulatory documentation, validation, and acceptance. MONAI carefully documents its operating environment, I/O methods, random seeds, data sources, and more to aid in these reproducibility efforts. Similarly, rather than forming an alternative to PyTorch, MONAI builds upon and maintains compatibility with PyTorch. Rather than creating its own I/O system, it uses ITK. Rather than implementing every data transform from scratch, it uses libraries from DKFZ, KCL, ICL, ITK, and others. Communities are coming together, rather than competing, and the power of open source is growing more rapidly as a result.
- Interactive Visualization and Best Practices: MONAI comes bundled with multiple Jupyter Notebooks that exemplify best practices for deep learning in medical imaging. These notebooks explore common tasks and replicate results from top performing networks in several medical imaging grand challenges. Furthermore, we have paired MONAI with ITKWidgets that bring interactive visualization and annotation capabilities to Jupyter Labs and Notebooks. ITKWidgets enable deep learning inputs, intermediate values, and results to be visualized, explored, and optionally refined by researchers and clinical collaborators with ease. Such interactive visualization of inputs and intermediate results are crucial to avoid hours and days of wasted training time and to gain new insights into results.
MONAI Getting Started: A Jupyter Notebook
MONAI is now available as a standard Python package, and it is now an official component of the PyTorch ecosystem. MONAI can be installed on any standard Python platform via
pip install monai itk itkwidgets You can verify the MONAI installation and the versions of its components in a jupyter notebook, as follows:
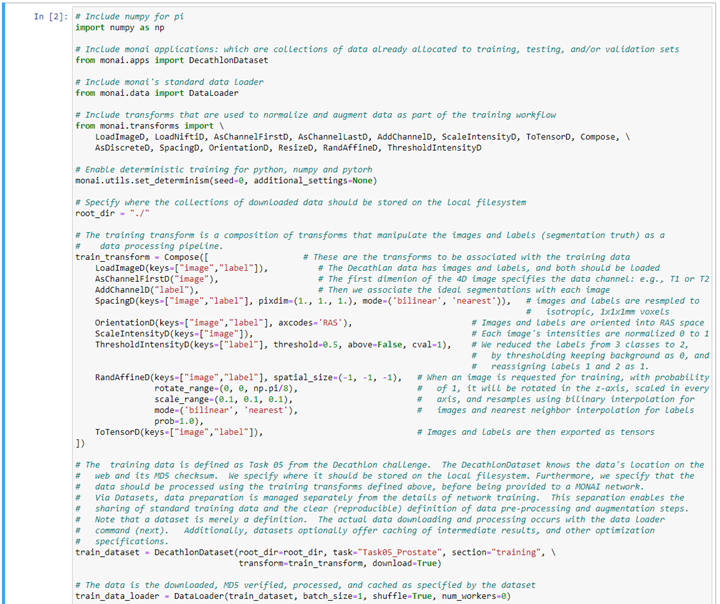
MONAI separates the specification of datasets from the specification of networks. In that way, a collection of training, validation, and testing data as well as a standard set of pre-processing and data augmentation steps for preparing that data can be clearly defined and shared with others, independent of the deep learning network that is then applied. This ability to share the specific data and preprocessing steps for an experiment / challenge is critical to reproducibility and rapidly advancing the field.
MONAI comes with multiple pre-defined “datasets.” These are collections of data on the web that have been pre-allocated into training, testing, and validation sets which can be downloaded and processed with a single MONAI command. We have also made it straightforward to add and share new MONAI datasets. Here we show setting up a training data pre-processing and augmentation workflow in MONAI:
Interactive visualization is critical to deep learning research, application development, and result review. We have paired MONAI with ITKWidgets to provide those visualizations, as demonstrated next:

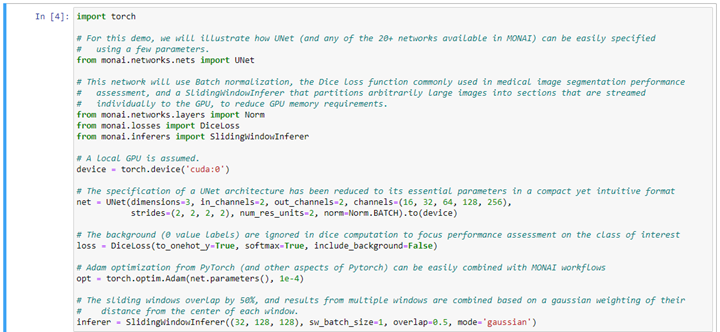
Once the training data of the network has been defined and reviewed, the network to be trained is specified. MONAI provides compact and intuitive parameterizations of the most popular and cutting-edge deep learning networks for medical imaging. It also provides training and inference engines that address GPU memory limitations and that enable multi-GPU training paradigms. Here is an example using a simple UNet definition:
With training data and networks defined, the training can be conducted and observed, as follows:
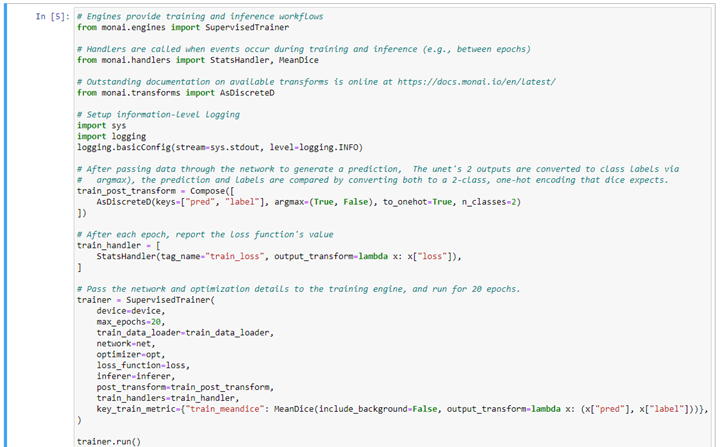
Once training is complete, the testing data from the challenge can be evaluated. Again, pre-processing steps (transforms) can be specified, but typically data augmentation methods are not applied to validation data. Results can be easily saved to disk in a variety of formats.
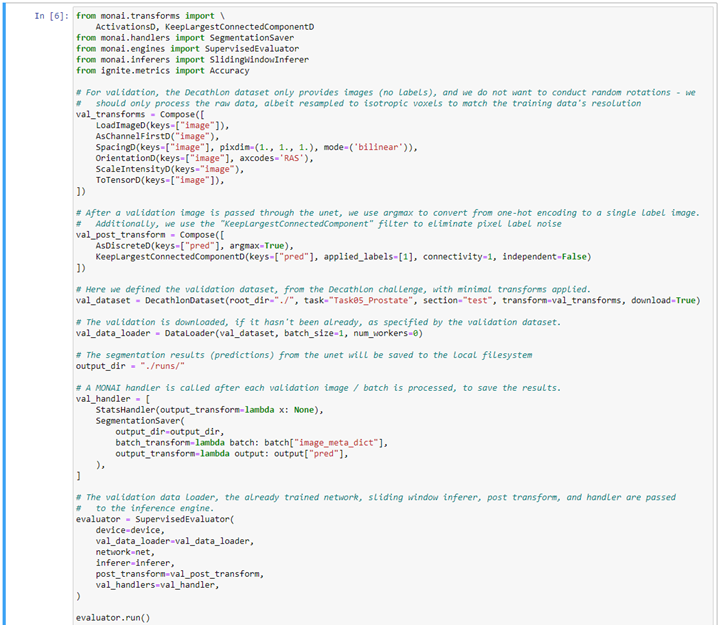
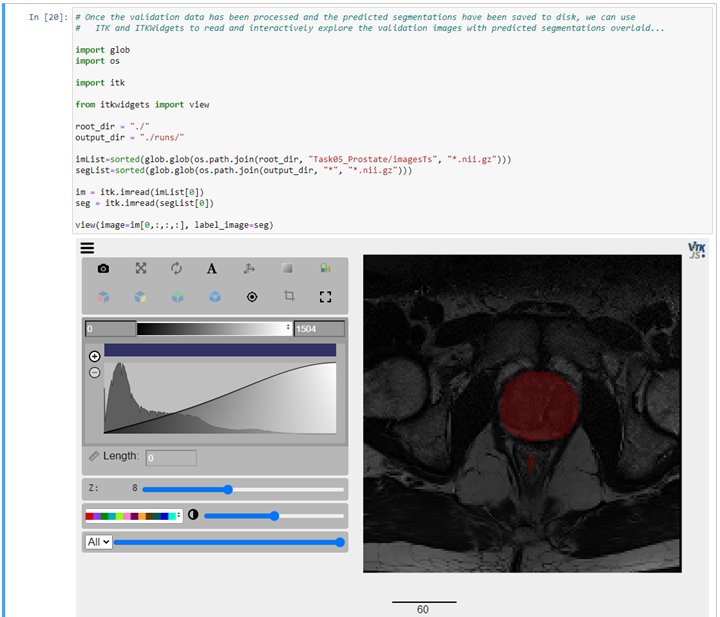
To view results, we recommend using ITKWidgets to generate both 2D (slice-based) as well as 3D renderings of the input data with the segmentation results overlaid. ITKWidgets also offers checkerboard and side-by-side comparison of images as well as the rendering of surfaces, meshes, point clouds, and many other data formats from ITK, numpy, and Pytorch/MONAI.
The instructions given in this section are available as a Jupyter Notebook on github at:
https://github.com/KitwareMedical/MONAI-GettingStarted/blob/main/MONAI-ITKWidgets.ipynb
Introductory Slides and Videos
We have also work with other members of the MONAI team to develop three sets of slides that are targeted for different audiences. These slides do provide some guidance on their own, but they are really meant to be used by lecturers who already know MONAI and who want to introduce MONAI to others.
1. MONAI: A three minute introduction:
https://www2.slideshare.net/StephenAylward1/monai-medical-image-deep-learning-a-3-minute-introduction
These slides were presented at the start of the 2020 3D Slicer Project Week. A video recording of that presentation is given here. Admittedly, the video contains some references specific to that venue, but the overall message is general and should help guide the use of the 3-minute introductory slides.
https://www.youtube.com/watch?v=tBrMVTlzb8s
2. MONAI for Data Scientists and Developers:
https://www2.slideshare.net/StephenAylward1/monai-medical-imaging-ai-for-data-scientists-and-developers-3d-slicer-project-week-2020
These slides were part of an hour long presentation on MONAI given during the 2020 3D Slicer Project Week. This presentation is geared towards Pytorch / Python users, deep learning researchers, and data scientists who would like an overview of why MONAI should be used for medical imaging deep learning research and application development.
3. MONAI and Open Science for Clinicians and Project Managers:
https://www2.slideshare.net/StephenAylward1/monai-and-open-science-for-medical-imaging-deep-learning-sipaim-2020
These slides were given as a keynote presentation at the 16th International Symposium on Medical Information Processing and Analysis (SIPAIM 2020). These slides promote open science practices, acknowledge open science as a major reason for the success of deep learning, and introduce MONAI as an open science platform. A recording of the keynote presentation is also available online:
https://youtu.be/XHJcKheftrc
Additional MONAI Material and Forums
- Getting Started as a MONAI User:
- Getting Started as a MONAI Developer:
- Community Guide:
https://github.com/Project-MONAI/MONAI#community
- Contributing Guide:
https://github.com/Project-MONAI/MONAI#contributing - Suggestions for first contributions:
Look for the “Good First Issue” tag on the MONAI issue tracker: https://github.com/Project-MONAI/MONAI/labels/good%20first%20issue
- Community Guide:
- Q&A forums:
- PyTorch Forums. Tag @monai or see the MONAI user page.
https://discuss.pytorch.org/u/MONAI/ - Stack Overflow:
https://stackoverflow.com/questions/tagged/monai - To join the MONAI Slack Channel, please fill out the Google form:
https://forms.gle/QTxJq3hFictp31UM9
- PyTorch Forums. Tag @monai or see the MONAI user page.
- MONAI News:
We are extremely excited about what MONAI has to offer and how well it integrates with existing standards and contributes to open science. If you need help introducing MONAI to your collaborators or integrating MONAI into your projects and processes, or if you have suggestions on improving MONAI, you may also contact us at kitware@kitware.com.
Thank you for this interesting post.
Can I read out the properties of the itkwidgets viewer object which I set manually in the UI?
When I create a viewer=view(…) object, change the gradient opacity in the UI, and query the value in the next cell (viewer.gradient_opacity), the property hasn’t changed.
I would like to dump the imaging device-specific adjustments in order to play them back the next time.