The Next Generation of Skeletal Representations in 3D Slicer
Kitware is excited to announce major updates to the SlicerSkeletalRepresentation extension, which revamps using, displaying, and storing skeletal representations (s-reps). These updates were developed as part of SlicerSALT for easier use and reuse of skeletal representations.
What is an s-rep?
S-reps are skeletal models that have been used for representing the shape of a wide variety of objects. They consist of a set of points sampled from a skeleton of the object together with vectors, called spokes, going from these samples to the object’s boundary. S-reps are constrained to be “as medial as possible” but offer additional flexibility that strictly medial models do not, which is particularly important for performing statistical analysis.
What’s new in this extension?
The SRep module
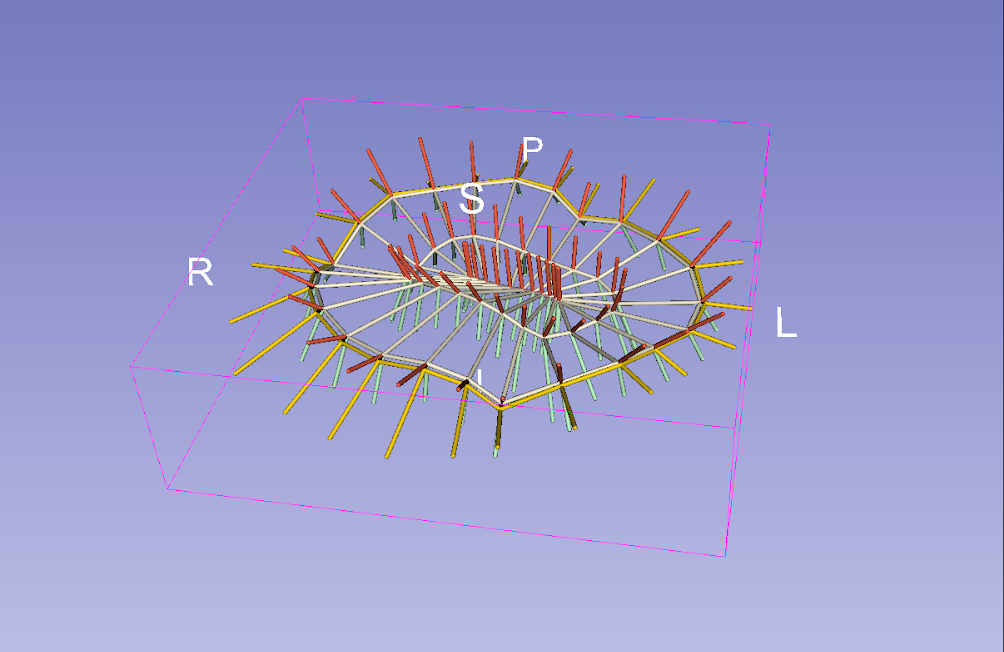
SRep is a new module that allows users to see basic information about s-reps, as well as modify their display properties. The user can easily change spoke and skeletal grid thicknesses, colors, opacity, and visibility for different parts of the s-rep.
This module also adds a new Medical Reality Modeling Language (MRML) data node for s-reps. This allows modules in this and other extensions to easily use, manipulate, and display s-rep data. The MRML nodes are available for use in both C++ and Python modules, and an advanced tutorial for creating new MRML nodes can be found on the 3D Slicer readthedocs.
An s-rep displayed with the new SRep module
A new .srep.json file type was added for saving and loading s-reps. This new file format has some advantages over the old xml+vtp file format previously used, including being more human readable and easier to copy to different machines (the old format had hard-coded, absolute file paths). These files can be read and written via the standard Slicer Save and Load buttons, as well as loaded via drag and drop. Saving s-reps to Medical Reality Bundle (MRB) files is also supported.
As noted in a previous blog post, the ShapePopulationViewer and Population Analysis (aka ShapeVariationAnalyzer) modules support the old s-rep file format (xml+vtp). Support has been added to these modules for the new .srep.json file format as well.
Loading an s-rep from a .srep.json file
Interpolation of s-reps is available for interpolating new spokes within the s-rep. The algorithm is the same as used in the old SRepRefiner module, with the exception of a bug fix to correct interpolation of skeletal grid points.
Interpolating an s-rep. The original s-rep was given thicker lines for visualization purposes.
S-rep nodes can also be transformed via linear transformations in the Transforms module.
The SRepCreator module
The SRepCreator module replaces SRepInitializer. It uses the same algorithm to create an initial fit of an s-rep to a Model node. Options exist to provide output that shows stepwise how the s-rep was fitted.
An initial fit s-rep with the model it was created from. Initial fits can then be refined to adjust the spokes and the boundary representations.
The SRepRefinement module
Once an initial fit of an s-rep has been made, it can be improved using the SRepRefinement module. This module replaces SRepRefiner. The new module uses the same refinement algorithm as the previous one, but altered to use the new s-rep MRML nodes.
How do you use the new extension?
- The new s-rep functionality will be available in the SlicerSALT 4.0 release.
- The s-rep extension can be installed in Slicer from the Extension Manager.
- Source code for the extension can be found on Github.
License
The SlicerSkeletalRepresentation extension is distributed under the Apache license, while Slicer itself is distributed under the BSD-style Slicer License. Both are permissive licenses that support unrestricted commercial use, with attributions.
Kitware’s medical computing capabilities
The SlicerSALT suite was developed by Kitware’s Medical Computing Team, which is composed of computer scientists, software engineers, and imaging experts. Specializing in the medical and biomedical industries, they provide collaborative research, development, and technology integration services for research centers, universities, and commercial companies. While the suite of tools is open source, you can partner with Kitware to ensure that you are effectively leveraging the technology. We can tailor the applications to meet your specific needs, including developing custom workflows, user interfaces, and more. For more information about partnering with Kitware, send us a message at kitware@kitware.com. You can also visit our commercial projects page.
Acknowledgements
Research reported in this publication was supported by the National Institute Of Biomedical Imaging And Bioengineering of the National Institutes of Health under Award Number R56EB021391. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.