The Role of ITK in the Era of Deep Learning in Medical Imaging
Deep learning currently dominates the field of medical image analysis. From brain tumor detection to cartilage segmentation and lung nodule analysis, convolutional and transformer-based neural networks often take center stage. Yet, behind every high-performing AI model, there is a need for reliable image preprocessing, spatial normalization, and data handling libraries. This is where the Insight Toolkit (ITK) continues to play a vital role.
ITK is an open-source, cross-platform library that has been powering medical imaging research for more than two decades. Started in 1999 by the U.S. National Library of Medicine, ITK has grown into a mature and widely adopted toolkit with contributions from hundreds of developers worldwide. While many new deep learning libraries have emerged, ITK remains essential because it solves fundamental problems:
- Reading complex medical formats like DICOM and NIfTI
- Handling spatial metadata with geometric fidelity
- Correcting scanner artifacts and harmonizing intensity ranges
- Standardizing input data so neural networks can learn reliably
In AI workflows, ITK continues to play a crucial role in ensuring that input medical images are geometrically and physically accurate before they reach a GPU. Here are a few examples.
Standardizing Anatomical Orientation
Consider a deep learning pipeline that we built in collaboration with UNC for osteoarthritis research, where knee MRIs from thousands of patients must be analyzed for cartilage thinning. Neural networks are sensitive to the orientation of their inputs. If one dataset encodes knees in left–right orientation and another in posterior–inferior orientation, the model may learn misleading features or fail.
This is where ITK helped us simplify the challenge. By using the OrientImageFilter, we can automatically reorient images into a canonical atlas orientation before they are passed into a neural network:
oriented_image = itk.orient_image_filter(
input_image,
use_image_direction=True, # respect metadata
desired_coordinate_orientation=dicom_pir # match atlas orientation
)In the same processing step, we applied a left–right mirroring operation, ensuring that all right knees are flipped to match a left-knee atlas. What appears to be a trivial preprocessing step is actually a key enabler of deep learning: by reliably standardizing orientation, ITK removed variability in the data, allowing the neural network to focus on learning biological patterns rather than scanner artifacts.
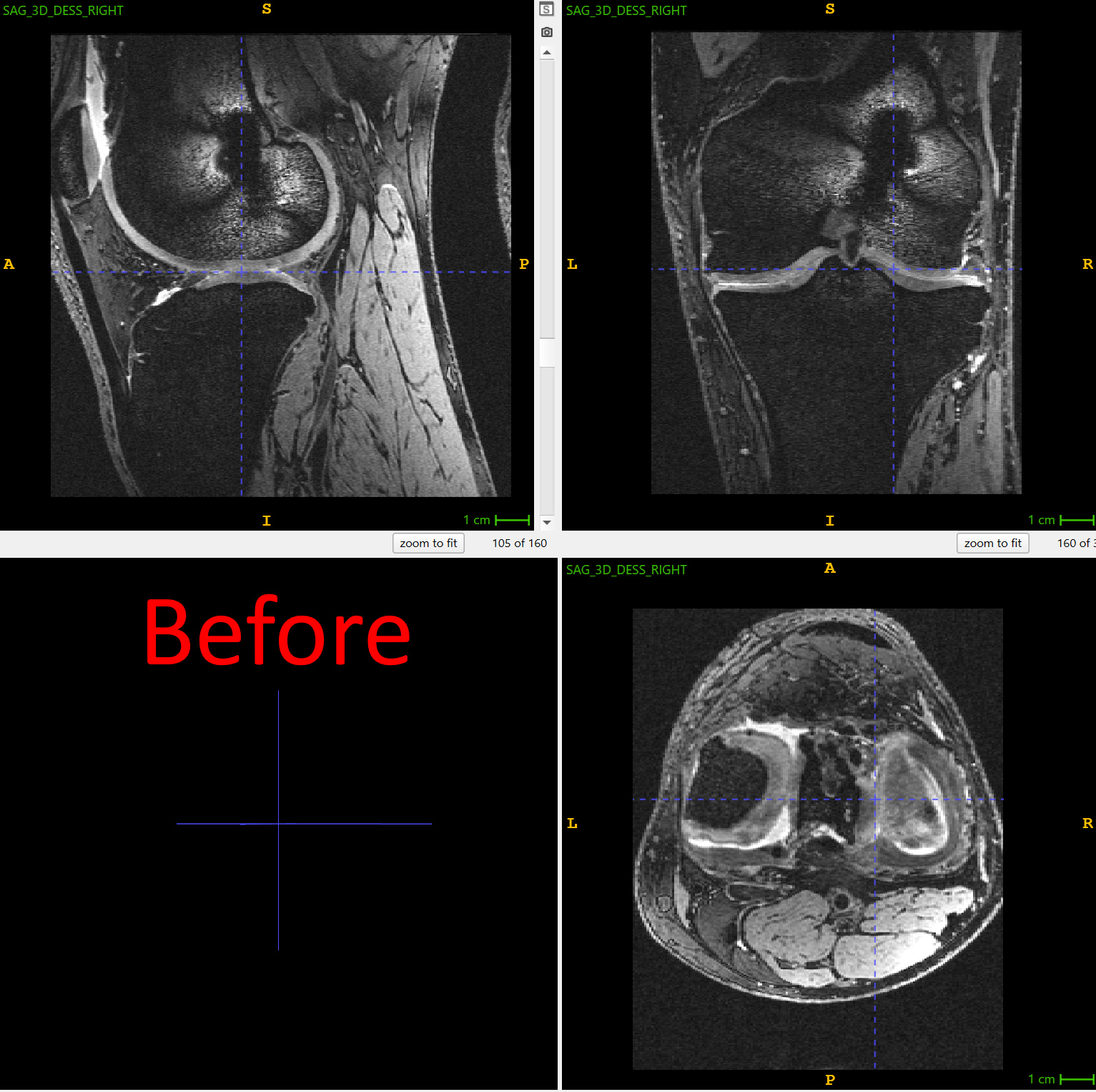
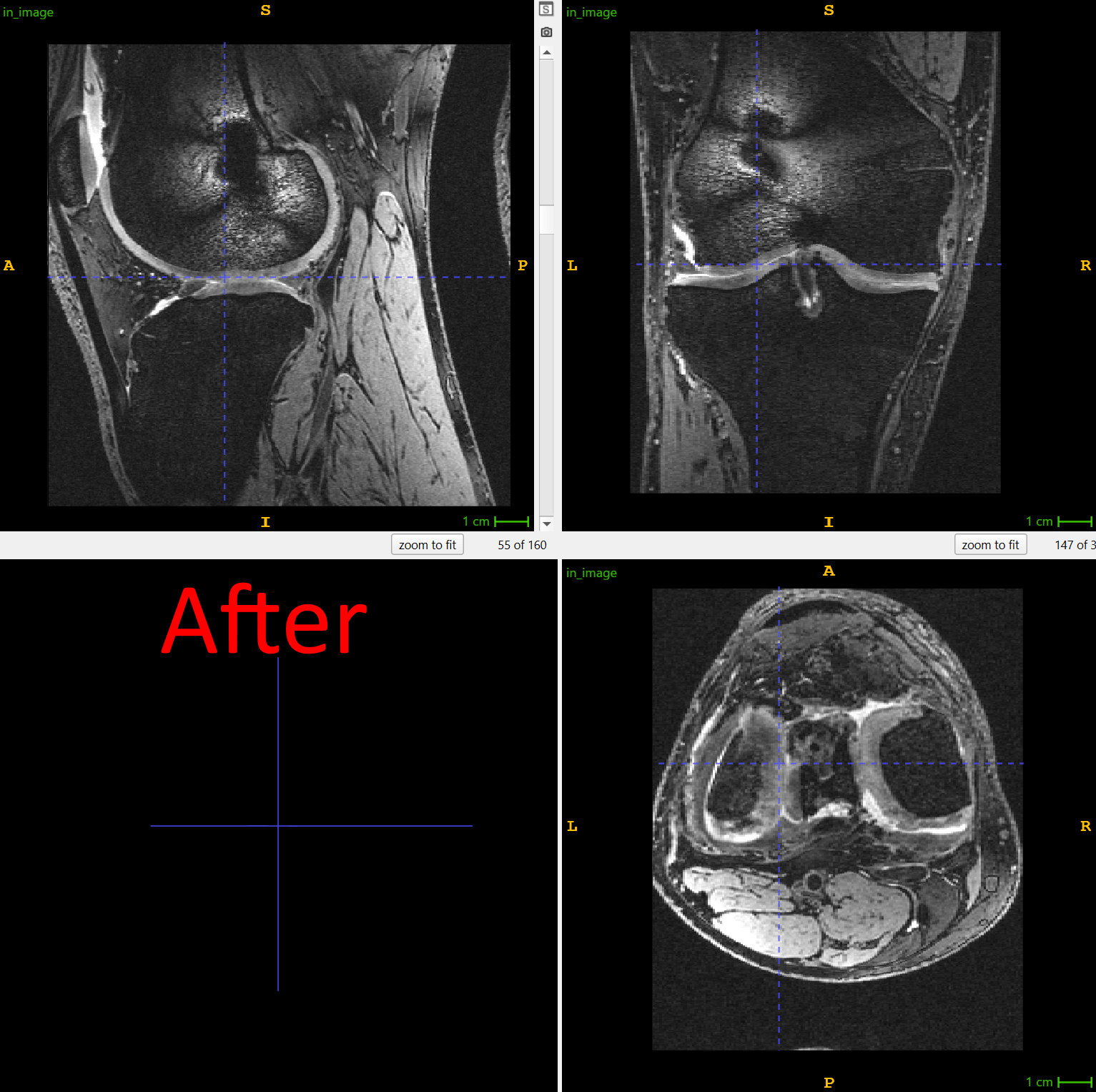
Figure 1: right knee is mirrored to match the atlas in the orientation of the left knees. Screenshots made using ITK-SNAP, one of the end-user applications built on top of ITK library. Use left-right slider to compare before vs. after.
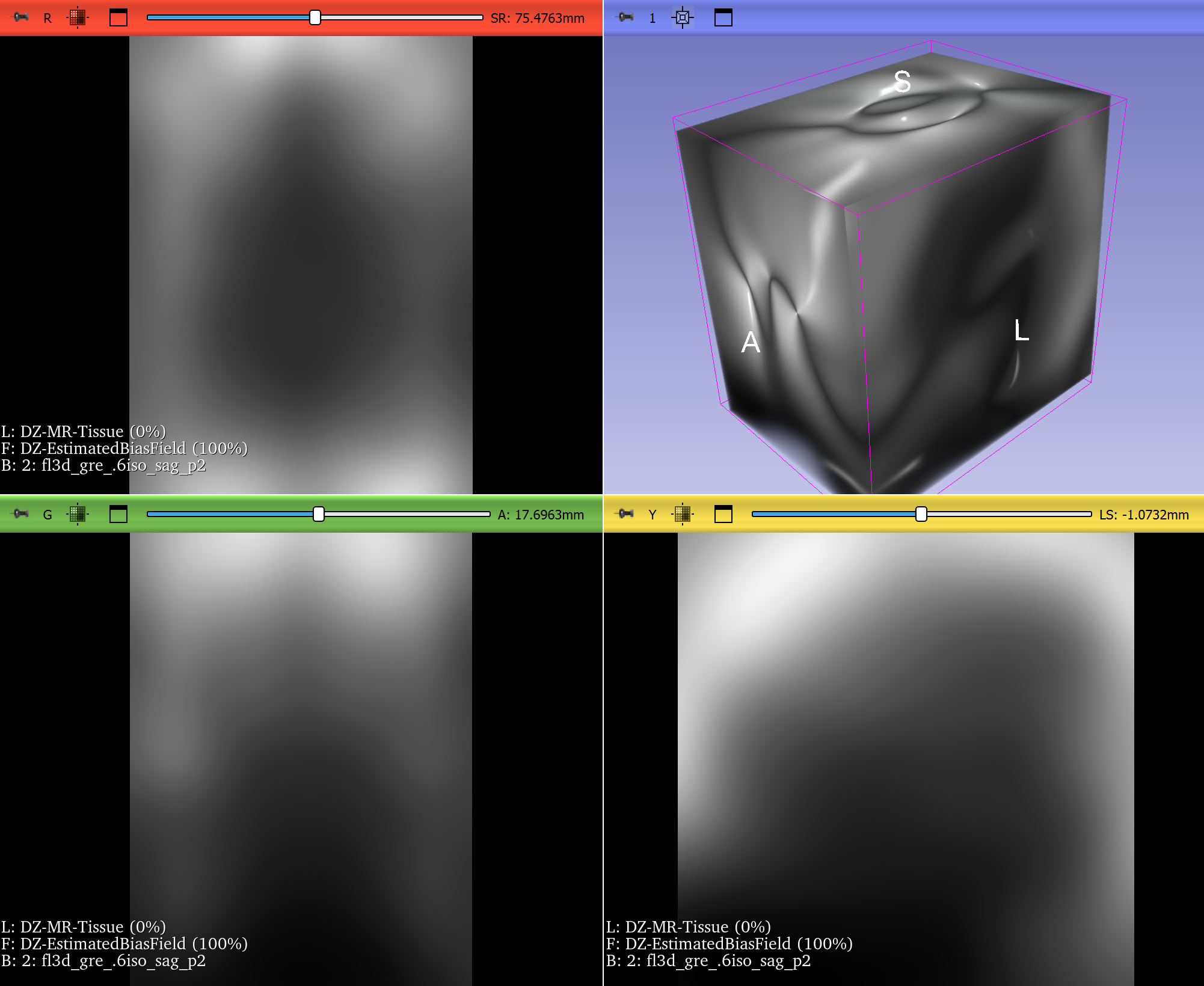
Bias Field Correction in Brain MRI
Magnetic resonance imaging (MRI) is prone to intensity inhomogeneities, also called bias fields. These low-frequency variations in intensity are not biological but caused by scanner physics. Deep learning models trained on uncorrected data risk learning scanner artifacts instead of tissue features.
ITK’s N4BiasFieldCorrectionImageFilter has become the gold standard for correcting this problem. With just a few lines of code, researchers can normalize the intensity profile of brain MRIs, making data consistent across scanners and institutions.
This correction is now a standard preprocessing step in brain tumor segmentation challenges and pipelines, ensuring that CNNs see consistent, biologically meaningful data. If a neural network is expected to encounter MRIs with intensity inhomogeneities, they can be intentionally introduced during training by using appropriate augmentation. Examples include RandBiasFieldd from MONAI and RandomBiasField from TorchIO. It can be even accomplished just by doing BiasFieldCorrection in reverse, starting from a corrected image and applying a randomly-generated bias field.
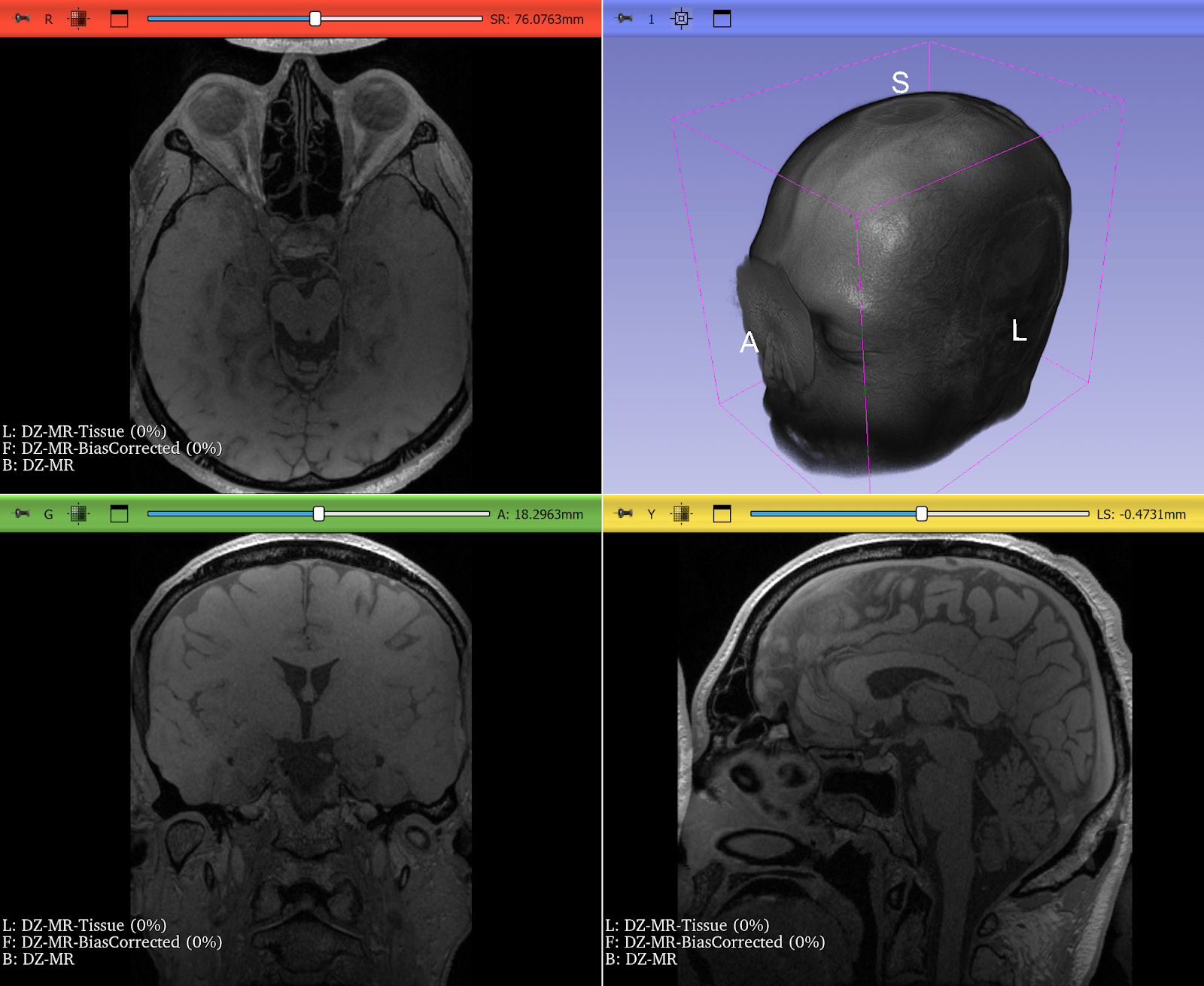
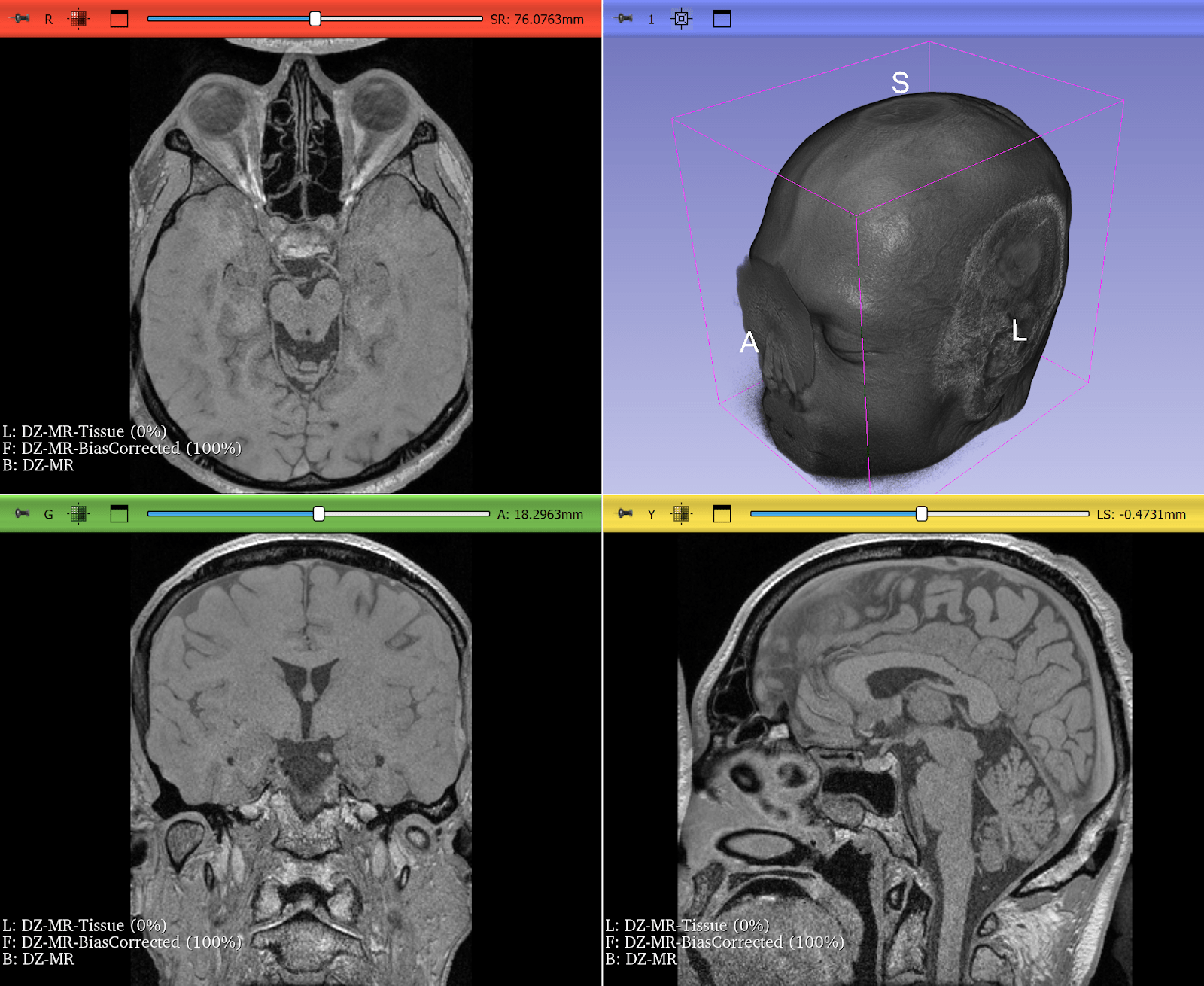
Figure 2: T2* Weighted MRI of the brain, before and after correction of intensity inhomogeneity. Bias field correction was done using 3D Slicer, another application prominently using ITK. Use left-right slider to compare before vs. after.
Resampling Lung CT Scans to Isotropic Voxels
Computed tomography (CT) scans often have anisotropic voxel spacing where the resolution along the axial slice direction is coarser than in-plane resolution. Deep learning models, especially 3D CNNs, perform poorly on such inconsistent geometry.
ITK provides robust resampling via the ResampleImageFilter, allowing researchers to convert CTs into isotropic voxel spacing (e.g., 1×1×1 mm³). This makes it quicker and easier to train deep learning models across heterogeneous datasets. In some cases, it can be as simple as:
resampled_image = itk.resample_image_filter(
input_image,
size=[256, 256, 256],
output_spacing=[1.0, 1.0, 1.0]
)This step is widely used in lung segmentation and COVID-19 CT analysis pipelines, where consistency in voxel geometry is critical for accurate volumetric measurements and model performance.
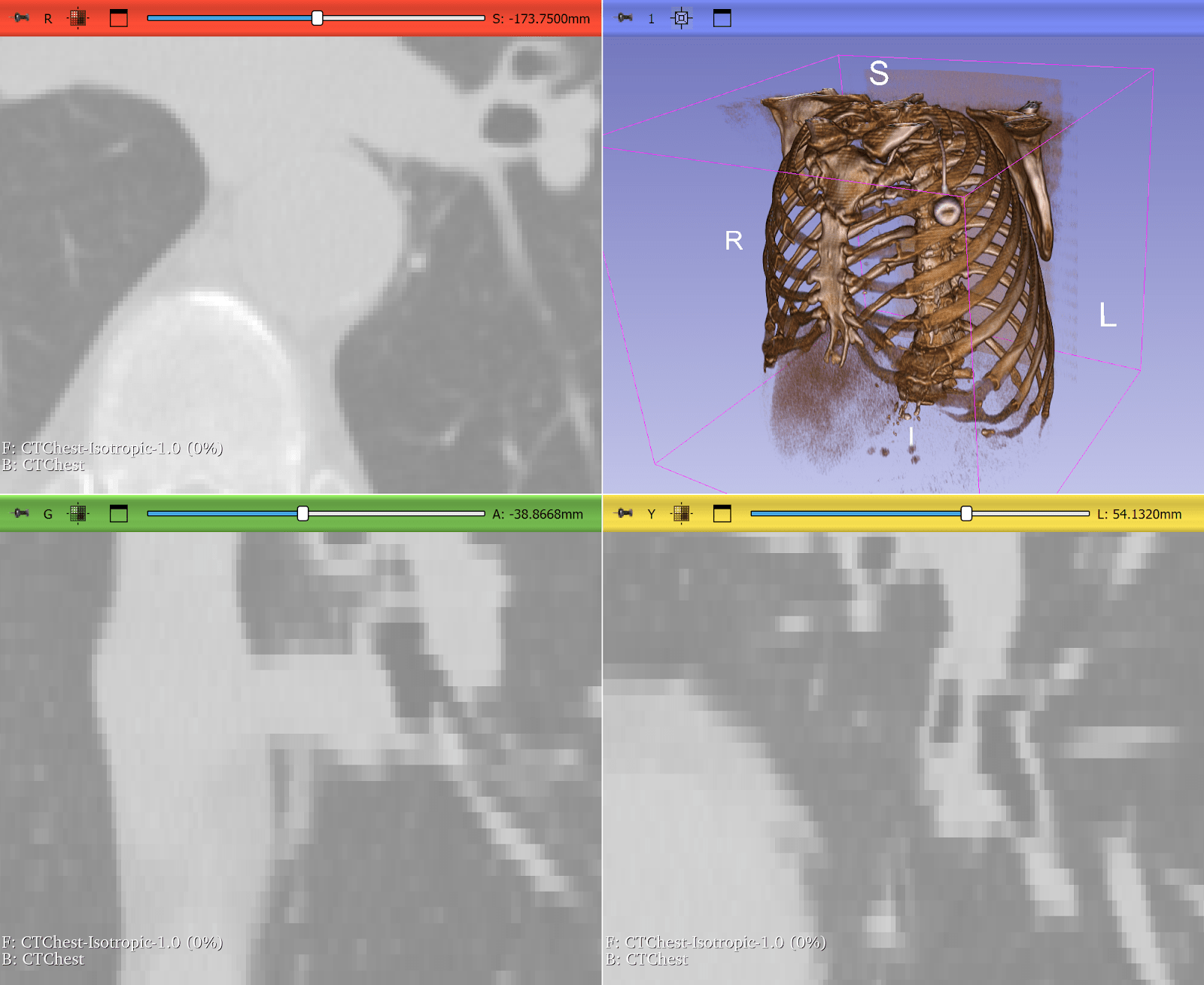
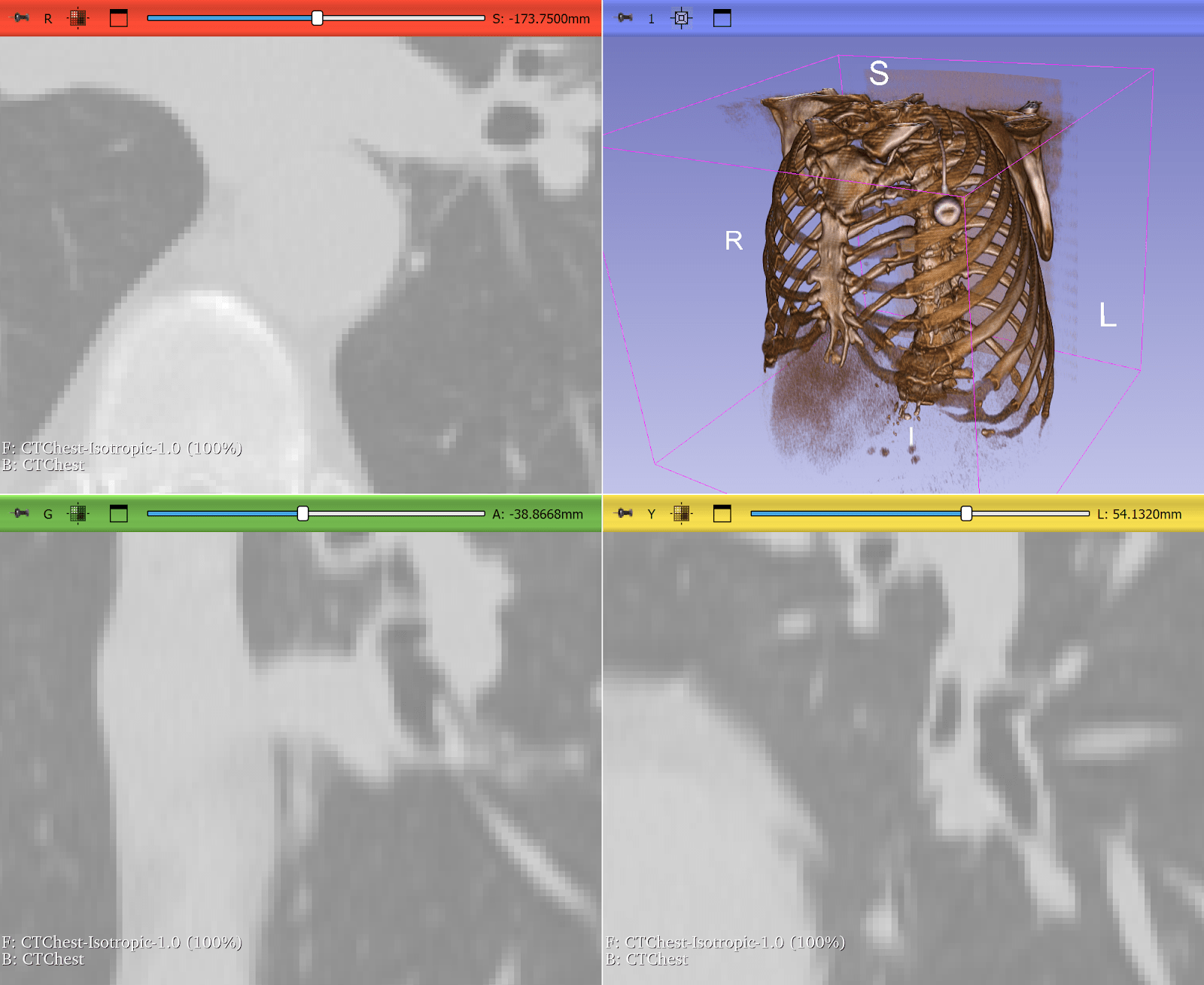
Figure 3: Lung CT before and after resampling, zoomed in with nearest neighbor resampling to show voxel sizes. Before: voxel size 0.76 x 0.76 x 2.5 mm. After: 1x1x1 mm. Use left-right slider to compare before vs. after.
Why ITK Still Matters in 2025
While deep learning frameworks like MONAI, PyTorch, and TensorFlow power the models, ITK remains a useful library that prepares the data. Its contributions fall into several key areas:
- I/O and metadata handling (DICOM, NIfTI, whole-slide pathology)
- Preprocessing and normalization (bias correction, intensity scaling, stain normalization)
- Geometric standardization (orientation, resampling, cropping, sometimes registration)
- Augmentation and transformations (elastic, affine, intensity-based)
- Interoperability (via SimpleITK, bridging into Python, R, Java, and deep learning frameworks)
Every high-performing AI pipeline in medical imaging, whether for research or clinical translation, depends on clean, consistent, and geometrically accurate data. ITK ensures exactly that.
Conclusion
While ITK is a rich reservoir of classical image analysis techniques that remain relevant for some medical imaging tasks with limited data, ITK also plays a critical role in today’s AI research. From reorienting knees in osteoarthritis studies, to correcting brain MRI bias fields, to resampling lung CTs for deep learning, ITK enables modern workflows to function reliably. As deep learning continues to evolve, ITK will continue to be maintained and supported to ensure medical images are ready for AI.
Partnering with Kitware for ITK Expertise
Kitware has been a core contributor to ITK since its inception, and we continue to maintain and extend the toolkit today. Our work ranges from advancing deep learning–based medical imaging workflows to developing statistical shape modeling frameworks built on ITK. If you are developing an ITK-based workflow or preparing imaging data for AI research, we welcome collaboration. Reach out to discuss how we can help design, optimize, or extend your pipeline with ITK.