Viime-Extract: AI for biomedical discovery from the scientific literature
Biomedical research today is generating increasingly vast and complex datasets along with an explosion of scientific literature that is overwhelming scientists’ ability to keep up. For example, PubMed is a free-to-access flagship scientific literature database maintained by NIH that has been growing exponentially for over 50 years (exceeding one million new publications per year over the last five years). In addition, new scientific databases and ontologies such as GEO, Reactome, KEGG, and HMDB are rapidly expanding. This accelerating research landscape requires new tools to assist researchers in understanding the many new types of data, study designs, questions, and novel biology so they can continue to make scientific breakthroughs at those same scales.
The true challenge in modern biomedical research is no longer access to knowledge, but how to retrieve and connect it together in meaningful ways in the face of seemingly unmanageable scale. The need for smarter tools to better extract new and actionable biomedical insights from the tidal wave of publications and data has never been greater.
In our Phase I SBIR project, Viime-Extract: An AI-powered Knowledge Graph Extraction System for Metabolomics, our mission was to help researchers turn this biomedical complexity into clarity. We accomplished this by leveraging AI technologies, specifically large language models (LLMs), to extract biomedical entities (such as genes, proteins, metabolites, and drugs) from scientific literature and link them with other databases and ontologies to create information-rich knowledge graphs. These graphs contain rich models of biomedical knowledge that researchers can query to draw connections between these entities to develop new biomedical insights. Additionally, they can leverage AI again to create new connections within and between knowledge graphs to suggest new interpretations of data and help generate new scientific hypotheses.
Viime-Extract: LLM-based Entity Extraction from Text
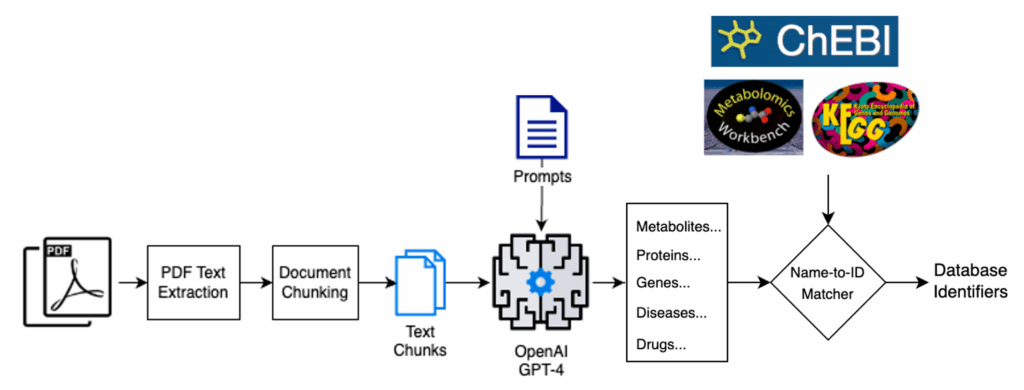
Viime-Extract is an open-source, Python-based workflow that leverages OpenAI’s GPT-4 language model to extract biomedical entities (metabolites, genes, proteins, drugs, diseases, and metabolic pathways) from scientific literature, connecting these through further processing to information from databases, resulting in a knowledge graph representing the information in the literature. Viime-Extract implements this specialized AI pipeline with the following features.
Text preprocessing and conditioning. The pipeline preprocesses text from the scientific literature, from abstracts to full papers, to create a suitable collection of input text chunks for subsequent processing by the LLM. This step prevents overflowing the LLM’s context window, and we take pains to overlap the text chunks to maintain as much continuity of context as possible.
Prompt engineering. Viime-Extract uses specialized prompt engineering to extract article metadata (title, authors, DOI), biomedical entities (metabolites, genes, proteins, drugs, diseases, and metabolic pathways), and the bibliography, yielding structured, contextually meaningful information ready for downstream integration. The prompts instruct the LLM that it is an expert in structured metabolomics information extraction and should extract the appropriate entities, taking care to be comprehensive, avoid errors, etc.
Fuzzy matching entity names. The system uses a fuzzy-matching disambiguator to resolve different but equivalent names to the same biochemical entity (e.g., “lactic acid”, “L-lactic acid”, and “lactate” all refer to the same chemical species). This step is essential for providing well-structured, clean outputs to the user, who would otherwise need to perform this disambiguation as an additional post-processing step. It also aids in searching the various databases for extra information about each entity.
Biomedical database connections. Viime-Extract aligns the output entities with leading biomedical databases, adding depth to each extracted entity by linking them with significant biomedical signatures such as gene functions, pathways, or metabolite properties. These, in turn, provide the richness in the resulting knowledge graphs, which now represent networks of biomedical entities as found and related in the literature search, augmented by high-quality information from community database resources.
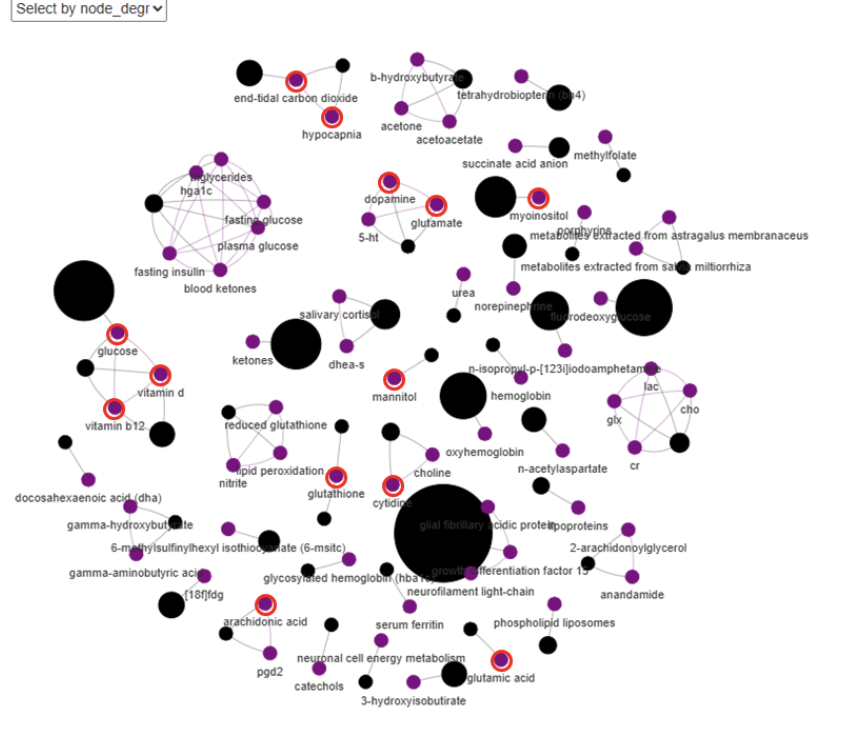
Interactive visualization and filtering. Finally, Viime-Extract visualizes the resulting knowledge graph consisting of the biomedical entities (as nodes) and their relationships (edges), both to each other and to information retrieved from public databases. These graphs can contain tens of thousands of nodes and edges, so a crucial feature of Viime-Extract is the ability to interactively filter, focus, and query these entities to address specific questions. Researchers can view the entire graph to see an overview of the encoded knowledge in a target set of papers, and then filter the graph interactively to focus on specific features of interest. For example, a knowledge graph might be filtered to show only the associations between a specific set of drugs, metabolites, and pathways to assist a researcher in studying a particular disease state. Another use case might leverage multiple knowledge graphs to compare the extracted gene signatures of two different diseases to study the question of whether they might share similar underlying biochemical mechanisms.
A New Lens on Science
Our work in this Phase I SBIR has demonstrated that LLM technologies, guided by carefully engineered prompts, can extract scientifically meaningful entities from research text with high accuracy. Furthermore, our domain-specific engineering efforts in text preparation, name disambiguation, database integration, and knowledge graph construction show that we can leverage the results of Viime-Extract to create richly connected, actionable knowledge graphs for researchers.
Viime-Extract offers researchers a powerful new way to understand biomedical literature. By linking insights from thousands of papers into interactive, filterable knowledge graphs, it enables scientists to quickly grasp complex research landscapes, identify new connections, and generate novel hypotheses. Moreover, Viime-Extract is a highly flexible and customizable workflow that can adapt to a wide range of scientific domains.
We are looking forward to continuing to develop Viime-Extract into a production-ready system that can scale to the changing needs of the biomedical research community. Our next steps include creating graph database systems to support efficient querying and advanced filtering of the knowledge graphs, scaling up the services to enable researchers at large to use the system, and adapting the Viime-Extract model to other scientific areas besides biomedical research.
Get In Touch!
If you are a scientist and you would like to accelerate the work of making sense of the vast research background so you can focus on your science, we want to hear from you! Please contact us for a demo of Viime-Extract and tell us about the specific problems you’re wrestling with. We are always interested in how we can engage with the scientific community to help solve problems with powerful tools like Viime-Extract.
This effort was an SBIR Contract Phase I funded by NIH/NIAID, carried out by Kitware and Indiana University, with a focus on metabolomics and infectious diseases.