Towards interactive selection with ParaView / LidarView
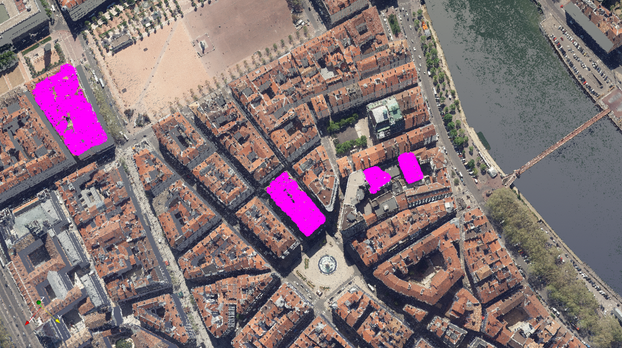
Interactive selection algorithms have always been an important aspect of any computer graphics and visualization software tools. Many software such as Blender, or AutoDesk include tools that facilitate the interaction with the scene and the edition of objects within it. ParaView recently introduced the selection editor, which helps its users select areas of interest with different selection algorithms (frustum-based, query-based etc…). Each selection instance can be composed with boolean operations. Although they can be very precise, we wanted to further improve its user-friendliness.
In this blog post, we’ll describe the developments of PV-SAM: our automatic selection plugin for ParaView/Lidarview that leverages Meta’s Segment Anything Model (SAM). We will also highlight several use-cases for different domains such as defense, industry and healthcare where this tool may be used.
Segment Anything Model for interactive segmentation
Interactive selection through segmentation is an active research topic. Not only does it help interact with data, but it is also useful for assisted dataset annotation in the context of computer vision (for ex. in manufacturing [1] or medical imaging [2]). This has gained a lot of interest since the AI-boom and the abundance of data.
One of the first approaches of segmentation was through active contour, or snake methods [3], which fit splines to edges of an image. Because they suffered from local minimas, the community moved towards global optimization methods such as graph cut [4], which partition an image represented as a graph into two disjoint subsets. Those methods can be really precise and robust, but are computationally expensive when transitioning to 3D, and require a lot of inputs from the user. More recent approaches rely on using unsupervised segmentation deep-leaning models, especially with transformers.
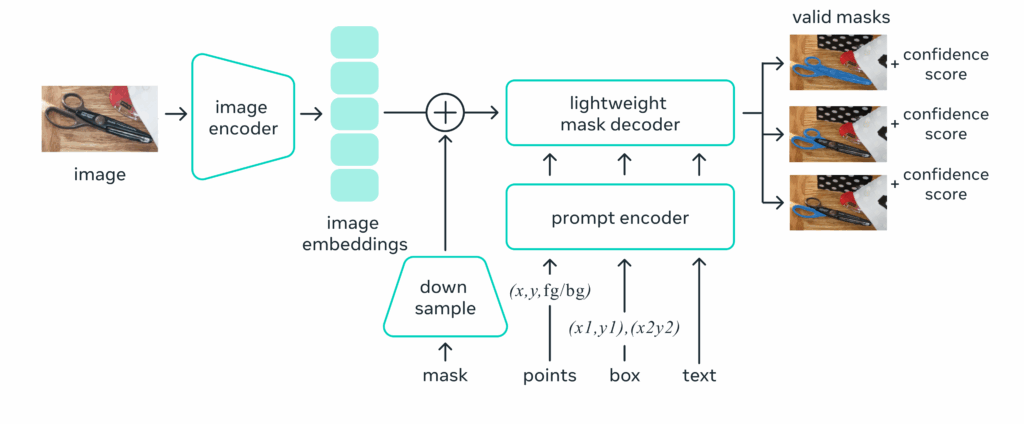
One of them gained a lot of attraction recently, Segment Anything (SAM) [5]. It enables efficient and accurate free-form segmentation of objects or regions within an image. From the user input (either a single click/point, a rectangle), the model will automatically segment the corresponding area in the image. SAM architecture consists of three main components:
- the image encoder: that extracts image characteristics using ViT [6]
- the prompt encoder: which transform user inputs into the same latent space as images
- the mask decoder: to decode information from the latent space and get the segmentation
The model is very efficient and flexible enough to handle multiple domains, however some limitations persist and require additional fine-tuning (for ex. in medical imaging [7]).
Inspired by projection-based methods from 3D detection, we can use the ParaView rendered image as a proxy for the scene. After selecting an area of interest with a single click on the rendered-view and calling SAM, we can get the associated cells on the scene by inverse-rendering. This makes our method powerful and agnostic to the underlying data.
VTK and ParaView plugin
SAM plugin
The SAM plugin can be broadly divided into two main components. The ParaView side handles two essential steps: capturing the rendered image together with the inference point, and applying the resulting selection back onto the 3D dataset. Between these two steps, the VTK-SAM component computes the segmentation mask from the rendered image and the user input. To keep the pipeline as simple and efficient as possible, we rely on existing ParaView/VTK utilities such as vtkWindowToImageFilter to convert the rendered view into a vtkImageData, and selectPolygonPoints, which allows selecting points on the rendered image from a polygon passed as an argument. The latter is particularly advantageous, since it enables VTK-SAM to directly leverage the full range of ParaView’s selection features, such as add, delete, toggle, grow selection, and many others.
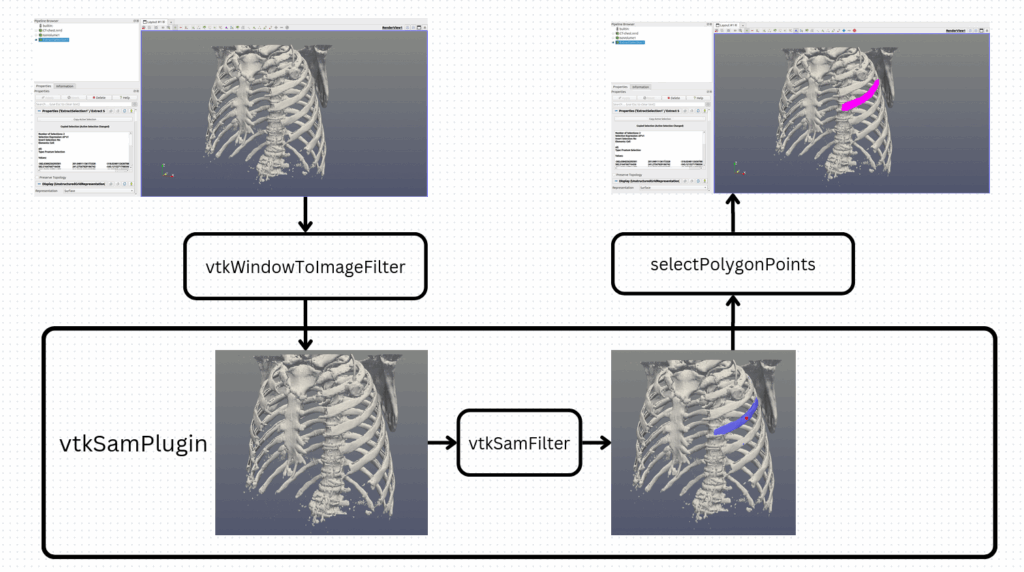
Figure 2. PV-SAM plugin overview
VTK SAM
VTK-SAM represents the VTK filter part of the plugin. It is composed of two core filters: vtkSamEncoder and vtkSamInferer. The first step in developing these filters was to export the SAM models to ONNX using a dedicated exporter specifically implemented for SAM. Once exported, both the encoder and the inferer can be executed with the ONNX Runtime library in C++, and then wrapped inside VTK filters.
The vtkSamEncoder filter preprocesses an image and calls the ViT encoder (provided as an ONNX file). Its output embeddings are then passed to vtkSamInferer, along with the input point and the original image, to generate three masks. These masks can then be applied to an image and visualized with a third filter, vtkSamViewer. For ease of use, this entire pipeline is wrapped into a higher-level filter, vtkSamFilter. To optimize performance, vtkSamEncoder maintains a hash of the last processed image so that the encoding step is only recomputed if the image changes. Since image encoding is the most computationally expensive step, this significantly reduces runtime.
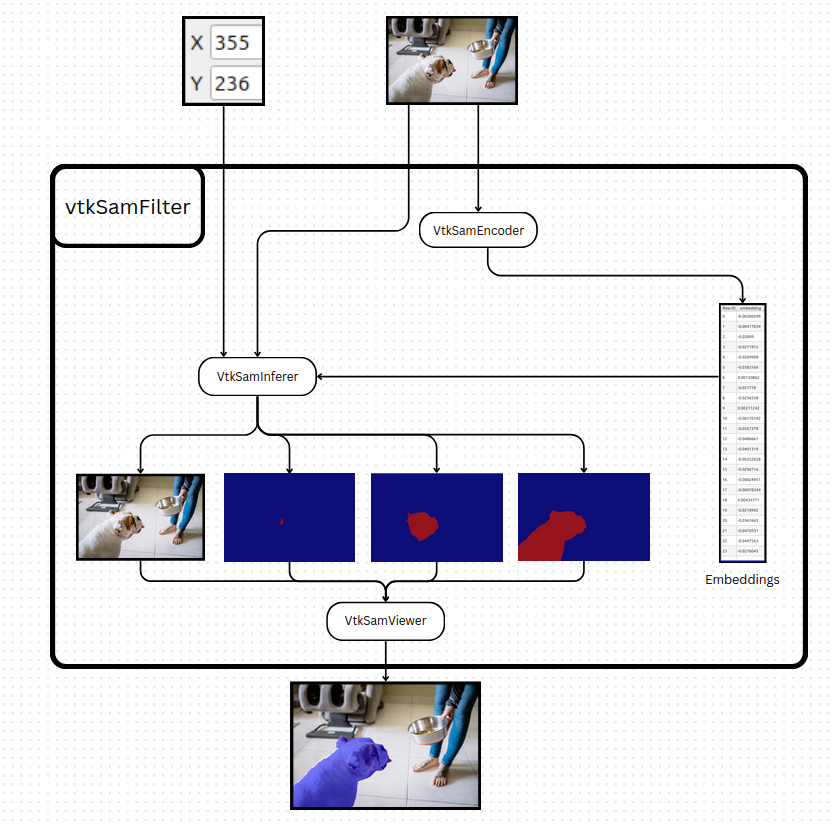
Figure 3. Vtk Sam overview
Plugin interface
The SAM plugin integrates seamlessly with ParaView’s native selection tools. It is available directly from the selection toolbar, making it possible to combine SAM-based segmentation with existing selection features. To use it, simply activate the tool and double-click on the region of interest in the dataset. The resulting mask can then be refined or combined with other selection operations, just like any other ParaView selection.

Results
We will now highlight some practical use-cases for the PV-SAM plugin.
Use-case 1: Remote sensing LIDAR
A first example is segmenting a point cloud from the remote sensing ahn4 dataset. We used a Calculator filter to rescale the RGB texture data (from uint16 to uint8) from the input LAS point cloud, with the following formula:
(255-Color_X * iHat * 255/65280)+(255-Color_Y * jHat * 255/65280)+(255-Color_Z * kHat * 255/65280)Then, we updated the scalar coloring attribute so it maps the RGB colors correctly, by un-checking Map Scalars under the Scalar Coloring property section.
Use-case 2: Industrial inspection
Here, we show how we can leverage this plugin to segment parts of infrastructure, this time for automated rail inspection. The SNCF data was downloaded from their open-data platform.
Use-case 3: Segmentation for SfM pipelines
Using the vtkSAM filter instead of PV-SAM, we can process on the image data directly (not the rendered view). In the following example, we are using Tsukuba data from the middleblurry stereo dataset.
Use-case 4: Whole body CT segmentation
Another use-case is a segmentation of bones from the left rib-cage. The surface was extracted after applying an iso-surface filter from the original cardiac CT volume available in slicer data samples.
It is important to understand that this is using the base original ViT model. For more accurate and robust results, especially if you are using the imaging data directly (instead of the surface model), you are encouraged to use a fine-tuned version of ViT like MedSAM for medical imaging [7].
Use-case 5: Volumetric environmental data
As a final example, we applied PV-SAM to a volumetric dataset of hurricane Isabel. After extracting a surface with the Contour filter, SAM could be used directly on the rendered geometry to isolate the cyclone’s core. With just a few simple interactions and the grow selection feature, it was possible to obtain a mask highlighting the rotating central region, making the complex structure of the storm much easier to visualize and analyze.
Data acknowledgement : Weather Research and Forecast (WRF) model, courtesy of NCAR and the U.S. National Science Foundation (NSF).URL : https://www.earthsystemgrid.org/dataset/isabeldata.html
Contact
And you, what would you do with this plugin ?
Check also our other topics around LiDAR processing with LidarView, and our blog post on ParaLabel, the integration of PV-SAM in it is on its way!
Contact us if you have an application in mind that we could help you build!
References
[1] Zhou, Longfei, Lin Zhang, and Nicholas Konz. “Computer vision techniques in manufacturing.” IEEE Transactions on Systems, Man, and Cybernetics: Systems 53.1 (2022): 105-117.
[2] Diaz-Pinto, Andres, et al. “Monai label: A framework for ai-assisted interactive labeling of 3d medical images.” Medical Image Analysis 95 (2024): 103207.
[3] Kass, Michael, Andrew Witkin, and Demetri Terzopoulos. “Snakes: Active contour models.” International journal of computer vision 1.4 (1988): 321-331.
[4] Boykov, Yuri Y., and M-P. Jolly. “Interactive graph cuts for optimal boundary & region segmentation of objects in ND images.” Proceedings eighth IEEE international conference on computer vision. ICCV 2001. Vol. 1. IEEE, 2001.
[5] Kirillov, Alexander, et al. “Segment anything.” Proceedings of the IEEE/CVF international conference on computer vision. 2023.
[6] Dosovitskiy, Alexey, et al. “An image is worth 16×16 words: Transformers for image recognition at scale.” arXiv preprint arXiv:2010.11929 (2020).
[7] Ma, Jun, et al. “Segment anything in medical images.” Nature Communications 15.1 (2024): 654.