Recap of the 44th NA-MIC Project Week in Gran Canaria

The 44th NA-MIC Project Week took place January 26-30, 2026 in Las Palmas de Gran Canaria, Spain. This marked 21 years since the first Project Week was held in Salt Lake City in 2005, and the community continues to grow and evolve. This year’s event brought together researchers, developers, and clinical professionals from around the world to collaborate on advancing open-source medical image computing software, with a particular focus on 3D Slicer.

Event Overview
Project Week 44 hosted 49 projects spanning eight categories: Infrastructure, Segmentation/Classification/Landmarking, Quantification and Computation, VR/AR and Rendering, Cloud/Web, IGT and Training, DICOM, and AI. The event drew participants from institutions across Europe, North America, and beyond, united by their work with 3D Slicer and the broader open-source medical imaging ecosystem.
A Year Marked by AI and Agent-Based Development
Walking around the room at Project Week 44, you’d notice something new: a lot of people were coding with AI assistants. Many participants were using tools like Claude Code to accelerate their development, and several projects explored how LLMs can assist researchers in querying data, running analysis pipelines, and even controlling imaging software with natural language.
Claude Skills for Imaging Data Commons: Andrey Fedorov’s team developed Claude AI skills for the NCI Imaging Data Commons, enabling researchers to query medical imaging datasets using natural language. The skill allows users to ask questions like “How many CT scans does NLST have?” and get accurate, programmatically retrieved answers.
MHub.ai MCP Server: Leonard Nürnberg developed a proof-of-concept MHubSkill that enables AI agents to discover, run, and adapt AI models from the MHub.ai repository. This work opens the door to automatic model selection and execution based on natural language requests.
Talk2View: The Talk2View team presented their natural language interface for medical imaging software, allowing users to control 3D Slicer using conversational commands. This project exemplifies the growing interest in making complex medical imaging tools more accessible through AI-mediated interactions.
LLM Vision for Medical Imaging: Michael Halle developed tools to help LLMs “see” medical images from IDC, bridging the gap between text-based AI assistants and visual medical data analysis.
Kitware’s Contributions
Three Kitware team members participated in Project Week 44: Sam Horvath, Ebrahim Ebrahim, and Thibault Pelletier. They contributed to several impactful projects. Sam Horvath also served on the global organizing committee.
SlicerOpenNav: Surgical Navigation for the Community
Sam Horvath led work on SlicerOpenNav, an effort to bring the functionality of NousNav (a Slicer custom application for surgical navigation) into a standard Slicer extension that the broader community can use and build upon. During Project Week, the team:
- Released NousNav 1.1.0 with usability improvements
- Created SlicerOpenNav by extracting and refactoring NousNav’s modules (preserving git history)
- Verified the extension works from build tree, installation, and source tree
- Refactored NousNav itself to depend on SlicerOpenNav, with the Home module remaining as a customization point
This work makes surgical navigation capabilities more accessible to the Slicer community while maintaining NousNav as a focused clinical application.
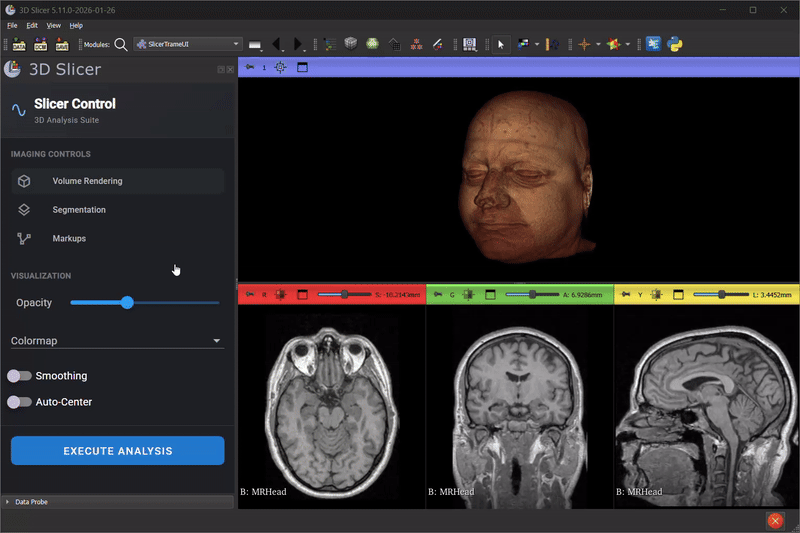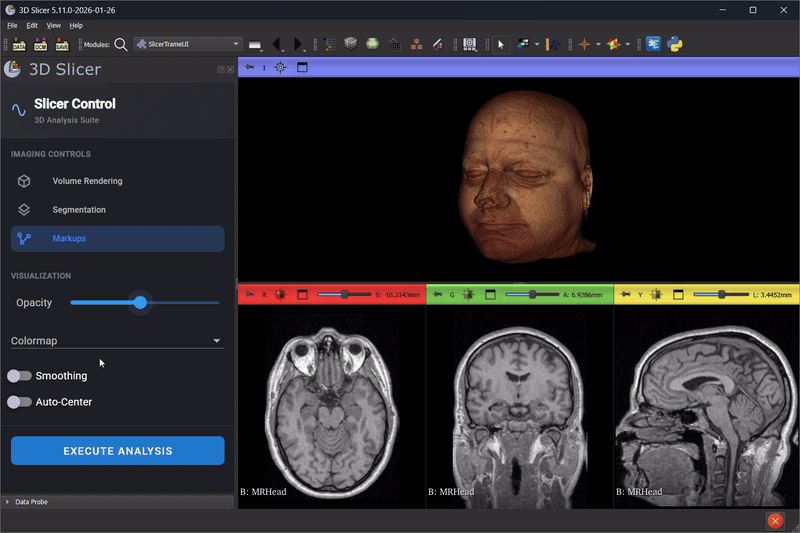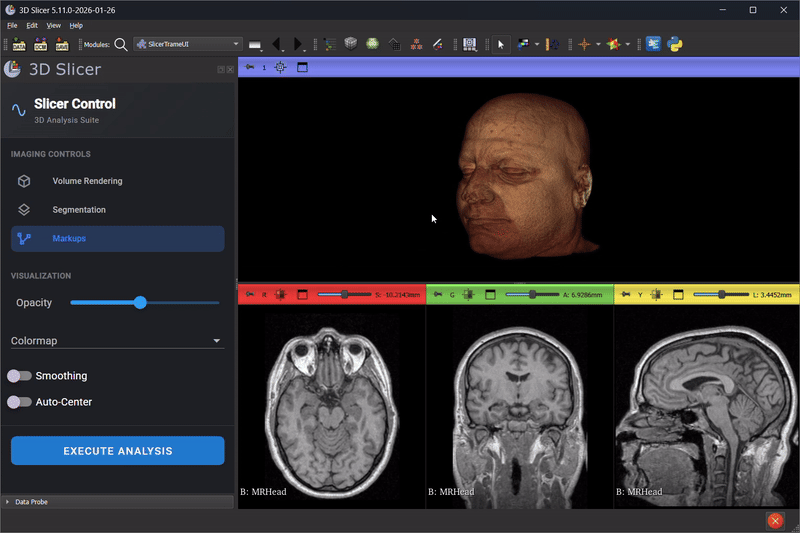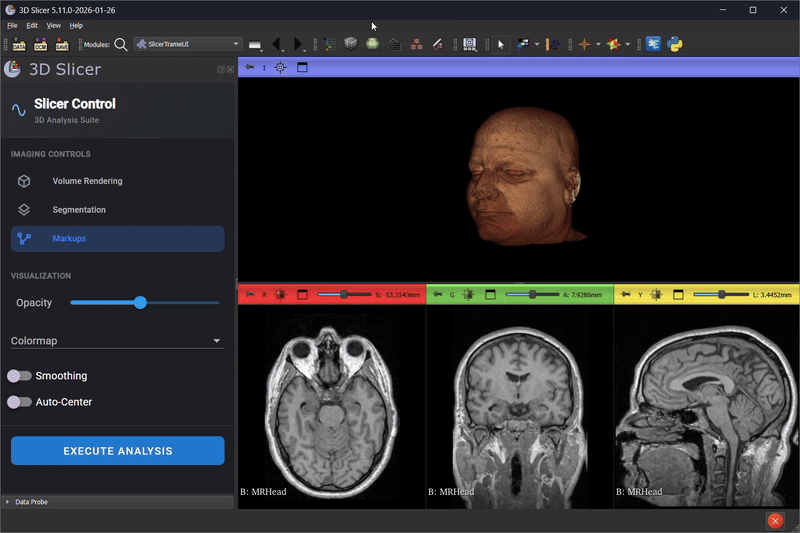
Scene Mirroring with trame-slicer
Thibault Pelletier continued advancing trame-slicer, working toward scene mirroring between 3D Slicer and web applications. Key achievements included:
- A proof-of-concept for running a trame-slicer server alongside a Qt Python desktop application using PyQt6 and qasync
- Testing approaches for non-blocking trame server integration with 3D Slicer to enable the integration of modern Web based UI in 3D Slicer desktop application
- Identifying technical blockers for Python Qt multi-threading in the 3D Slicer embedded Python environment
This work lays the foundation for future capabilities like real-time collaborative review, remote display on lightweight devices, and interactive teaching modules.
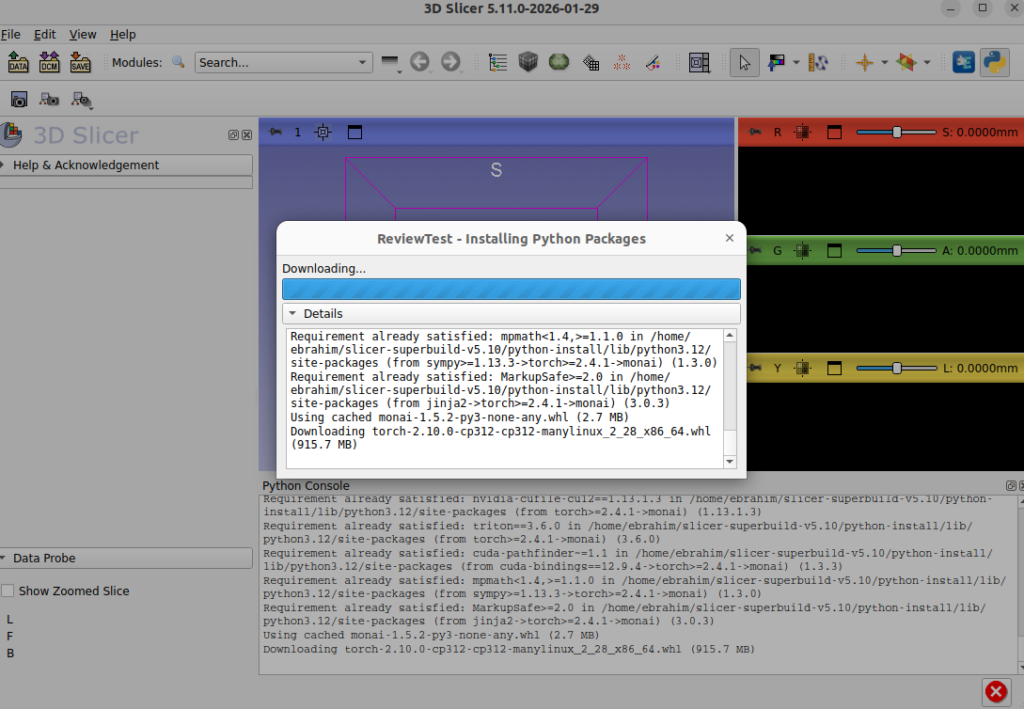
Python Dependencies Framework for Slicer Extensions
Ebrahim Ebrahim led an infrastructure initiative to improve how 3D Slicer extensions handle external Python dependencies. The project delivered:
- An analysis of the 94 currently indexed Slicer extensions that have external Python dependencies, and their dependency management patterns
- A pull request (Slicer#9010) introducing new utility functions (
load_requirements(),pip_check(),pip_ensure()) for dependency handling and extending the existingpip_install()utility. - Best practices documentation for extension developers
- Support for progress display during installation and prevention of blocking operations
This work addresses a long-standing pain point for extension developers and will make it significantly easier to build Slicer extensions that rely on external Python packages.
slicer.util.pip_installDMRI Population Analysis Library
Ebrahim Ebrahim explored bridging abcdmicro (a Python library for diffusion MRI population analysis) with 3D Slicer. The main outcome was adding example notebooks demonstrating NODDI estimation, tract segmentation, and multimodal population template construction—now with built-in example data so anyone can try them. Discussions with Arthur Chakwizira (BWH) helped identify future directions, and a proof-of-concept approach for the Slicer bridge was sketched out, though the bridge itself remains future work.
Project Highlights Across Categories
With 49 impressive projects spanning eight categories, we will only highlight a selection here. For the full list of projects and their outcomes, visit the PW44 project pages.
Infrastructure
Cast: Martin Bellehumeur extended FHIRcast for non-FHIR data, enabling multi-user collaborative workflows across healthcare applications including OHIF, 3D Slicer, and mobile devices. The project implemented bidirectional WebSocket communication and multi-user annotation synchronization.
The MHub.ai Slicer Extension (Leonard Nürnberg) underwent validation testing, with the goal of providing access to approximately 30 AI models directly within 3D Slicer via Docker containers. The team collected user feedback to improve the setup experience.
Segmentation and Classification
Using Automatic AI Segmentation Tools for IDC Data Enrichment (Lena Giebeler et al.) evaluated multiple segmentation models (MOOSE, CADS) on a representative subset of the NLST dataset and submitted CrossSegmentationExplorer for inclusion as a Tier 1 Slicer extension.
SlicerAdaptiveBrush (Ben Zwick, Andy Huynh) advanced their adaptive brush tool for semi-automatic segmentation, adding comprehensive documentation, performance caching, and preparing for Extension Index submission.
Quantification and Computation
DeformView (Isabel Frolick, Elise Donszelmann-Lund, D. Louis Collins) improved brain shift visualization during neurosurgery with dense displacement maps and Jacobian visualization. A user study showed 80% of participants preferred it over existing visualization tools.
Cloud and Web
Exploration of Foundation Models and Their Embeddings Using the Cloud (Deepa Krishnaswamy, Andrey Fedorov et al.) compared embeddings from 9 different foundation models on NLST lung CT data, building an interactive visualization tool for content-based image retrieval that helps identify similar patients.
VR/AR and Rendering
SlicerVirtualReality (Kyle Sunderland, Simon Drouin, Andras Lasso, Csaba Pinter) saw significant improvements: greatly improved markup rendering performance, confirmed that previously reported washed-out color issues are now resolved, and the extension has been re-listed on the Extension Index.
IGT and Training
Hands-on Tutorials Extension (Alejandro Rodríguez Moreno, Csaba Pinter) tackled the long-standing challenge of keeping Slicer tutorials up-to-date and accessible. The team began adapting an interactive tutorial framework from a commercial project for use in Slicer core. The framework guides users through steps using targeted tooltips and automatic validation, with support for tutorial dependencies and progress persistence—a more maintainable alternative to slide-based or video tutorials.
KidneyNav (Gabriella d’Albenzio, Kyle Sunderland et al.) demonstrated real-time kidney ultrasound segmentation and 3D reconstruction using an integrated nnU-Net model with OpenIGTLink streaming.
Breakout Sessions
In addition to project work, PW44 featured several breakout sessions that brought participants together to discuss cross-cutting topics:
- AI Tools: An informal community discussion where participants shared experiences, tips, and lessons learned from using AI coding agents
- 3D Slicer Q&A: An interactive session covering new features in Slicer 5.10, including Python 3.12 support, 5D NRRD file handling, the new Line Profile module, secure DICOM communication via TLS, and infrastructure updates preparing for Slicer 6.0. The 3D Slicer breakout slides are available.
- AR/VR/Rendering: Topics included the new Layer Displayable Manager for custom rendering pipelines, the VTK transition from OpenGL to WebGPU, future directions for Slicer’s rendering architecture, and an update on SlicerVirtualReality development plans for the Meta Quest 3
- Simulation: A survey of simulation tools (SlicerSOFA, SOFA, FEBio, Newton, GetFEM, SimNIBS), application areas including needle biopsy, ocular surgery, lung deflation, and fracture prediction, plus discussion of Slicer integration options for external simulation processes
Looking Forward
Project Week 44 demonstrated the continued vitality of the open-source medical imaging community and its enthusiasm for integrating emerging AI technologies. The trend toward AI-assisted development and natural language interfaces for complex imaging tools is likely to accelerate, and Kitware is well-positioned to contribute to this evolution through our work on trame, Slicer core infrastructure, surgical navigation tools, and domain-specific analysis libraries.
The next Project Week (PW45) will take place June 22-26, 2026 at MIT in Boston, USA. We look forward to continuing these collaborations and advancing open-source medical imaging together.
Acknowledgments
We thank the local organizing committee– Juan Ruiz-Alzola, Csaba Pinter, and the Ebatinca team– for hosting another successful event in Gran Canaria. We also thank the global organizing committee including Tina Kapur, Simon Drouin, Rafael Palomar, Theodore Aptekarev, and Sam Horvath for their continued stewardship of this valuable community gathering.