New titles and promotions at Kitware Europe
November 10, 2020
Over the last year, with a growing number of staff members, Kitware Europe has undertaken a vast Human Resources project aiming to structure its organization. The main goals of this long-term work are to harmonize job positions, design career paths, as well as recognize talents and achievements. On behalf on the entire management team, I […]

CMake 3.19.0-rc3 is ready for testing
November 6, 2020
The third CMake 3.19 release candidate!

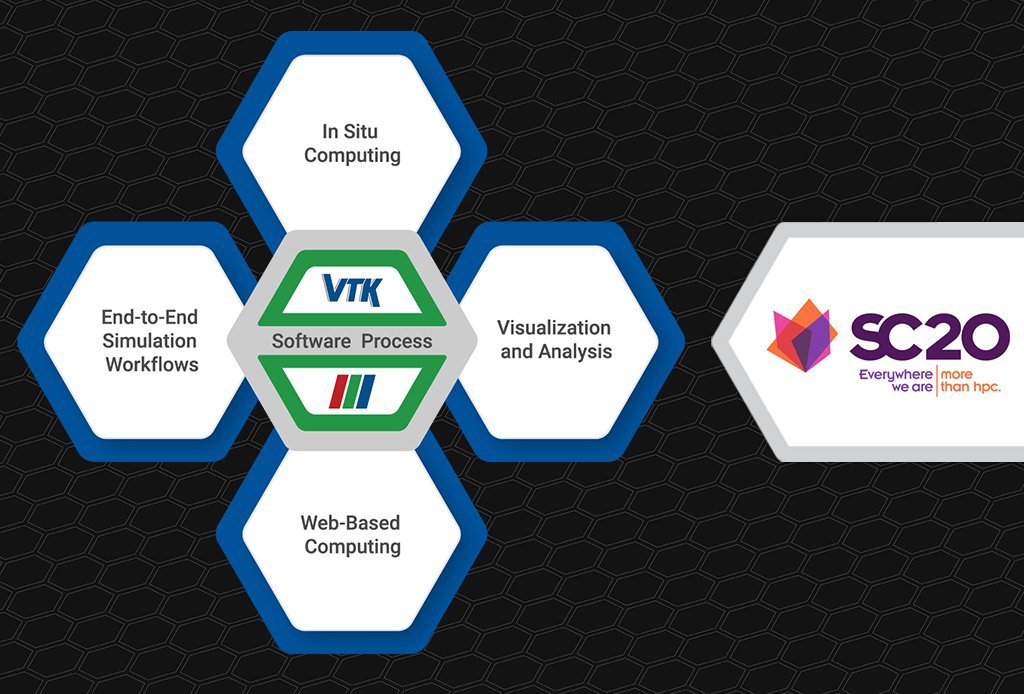
Kitware at SC20: Workshops, Presentations, and Software Releases
November 2, 2020
The International Conference for High Performance Computing Networking, Storage, and Analysis (SC20) brings together the international high performance computing (HPC) community. As scientists, engineers, researchers, educators, programmers, system administrators, and developers from around the world come together for the virtual conference this year, Kitware remains actively involved and looks forward to connecting with the HPC […]
CMake 3.19.0-rc2 is ready for testing
October 28, 2020
The second CMake 3.19 release candidate!

CMake 3.19.0-rc1 is ready for testing
October 13, 2020
The first CMake 3.19 release candidate!

CMake 3.18.4 available for download
October 6, 2020
We are pleased to announce that CMake 3.18.4 is now available for download

CMake 3.18.3 available for download
September 22, 2020
We are pleased to announce that CMake 3.18.3 is now available for download.

Integrating ITK and VTK: Give Us Your Feedback!
September 1, 2020
Please complete our online survey!

ITK 5.1.1 available for download
August 31, 2020
We are happy to announce the Insight Toolkit (ITK) 5.1.1! ITK is an open-source, cross-platform toolkit for N-dimensional scientific image processing, segmentation, and registration. Python Packages Install ITK Python packages with: or: Library Sources InsightToolkit-5.1.1.tar.gz InsightToolkit-5.1.1.zip Testing Data Unpack optional testing data in the same directory where the Library Source is unpacked. InsightData-5.1.1.tar.gz InsightData-5.1.1.zip Checksums […]

CMake 3.18.2 available for download
August 20, 2020
We are pleased to announce that CMake 3.18.2 is now available for download. Please use the latest release from our download page:https://cmake.org/download/ Thanks for your support! ---------------------------------------------------------------------------- Changes made since CMake 3.18.1: Alexandru Croitor (2): AutoGen: Fix over-specified direct dependencies of custom command AutoGen: Add test to check for correct AutoMoc dependencies Axel Huebl (1): […]

ParaView 5.8.1 Release Notes
August 5, 2020
ParaView 5.8.1 is a bug fix release that addresses 60 issues identified since the ParaView 5.8.0 release. The full list of resolved bugs with links to the bug reports is provided below. Readers and filters AMR Dual Clip and Contour Accept invalid input datasets Parallel Resample to Image pipeline doesn’t propagate all data Clip filter: […]

Pulse Python Support
July 22, 2020
On behalf of the Pulse Physiology Community, we are pleased to announce that Pulse now supports the Python programming language. Python developers can now create and control Pulse engines from any Python application. Binder Notebooks We have created a Binder based Jupyter notebook that creates a remote development environment in your browser to experiment with […]

Previous versions of ParaView supported volume rendering with independent mapping of colors and opacities to different components of a single input data array. However, it was available only for very specific dataset array formats, and it was not straightforward to create those arrays if they did not already exist. Coming in ParaView 5.9, ParaView’s user […]

CMake 3.18.0 available for download
July 15, 2020
I am happy to announce that CMake 3.18.0 is now available for download at: https://cmake.org/download/ Documentation is available at:https://cmake.org/cmake/help/v3.18 Release notes appear below and are also published athttps://cmake.org/cmake/help/v3.18/release/3.18.html Some of the more significant changes in CMake 3.18 are: The “CUDA” language can now be compiled using Clang on non-Windows platforms. Separable compilation is not yet […]

CMake 3.18.0-rc4 is ready for testing
July 10, 2020
I am proud to announce the fourth CMake 3.18 release candidate.https://cmake.org/download/ Documentation is available at:https://cmake.org/cmake/help/v3.18 Release notes appear below and are also published athttps://cmake.org/cmake/help/v3.18/release/3.18.html Some of the more significant changes in CMake 3.18 are: The “CUDA” language can now be compiled using Clang on non-Windows platforms. Separable compilation is not yet supported on any platform.This […]

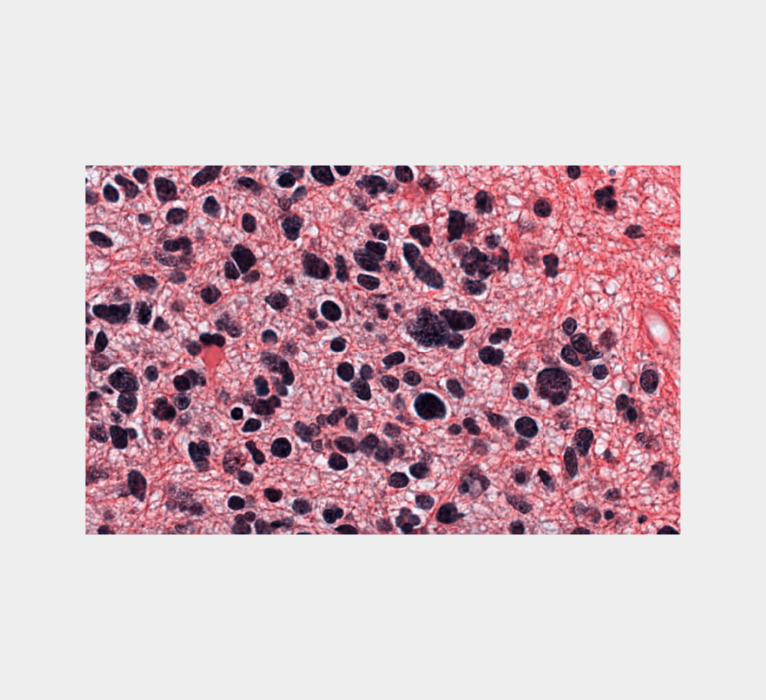
Introduction We have added a new module to the Insight Toolkit (ITK) to perform Structure Preserving Color Normalization on an H & E image using a reference image. The software is available in C++ and is also packaged for Python. H & E (hematoxylin and eosin) are stains used to color parts of cells in […]
CMake 3.18.0-rc3 is ready for testing
July 1, 2020
I am proud to announce the third CMake 3.18 release candidate.https://cmake.org/download/ Documentation is available at:https://cmake.org/cmake/help/v3.18 Release notes appear below and are also published athttps://cmake.org/cmake/help/v3.18/release/3.18.html Some of the more significant changes in CMake 3.18 are: The “CUDA” language can now be compiled using Clang on non-Windows platforms. Separable compilation is not yet supported on any platform.This […]

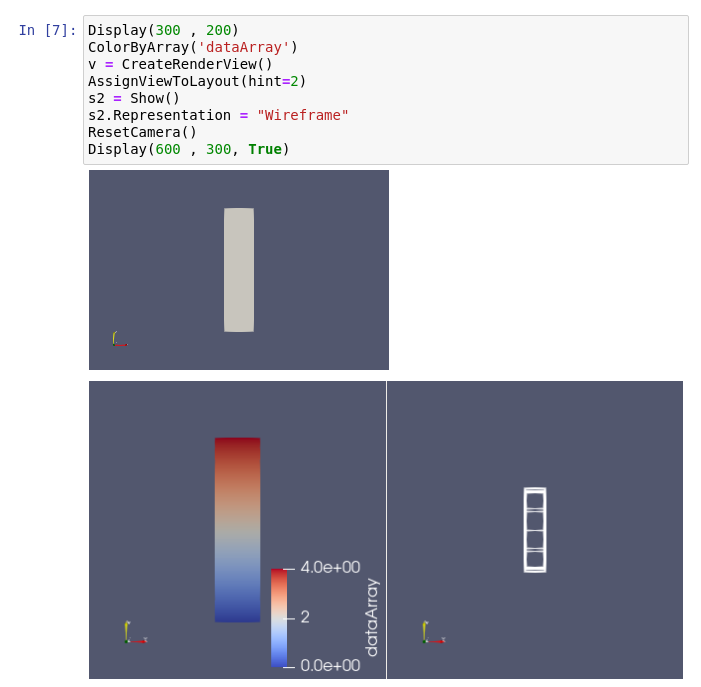
Controlling ParaView from Jupyter Notebook
June 30, 2020
We are happy to announce the release of a ParaView kernel for Jupyter notebooks [1] [2]! Basically, creating a ParaView Notebook launches the ParaView GUI in background. Then, the whole API of ParaView is available from the Notebook. At this point, it is very similar to the standard Python shell. Below is a demonstration video. […]